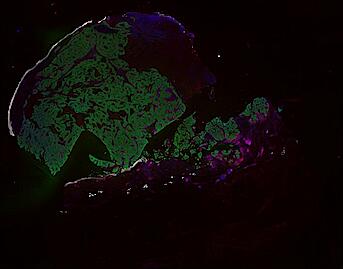
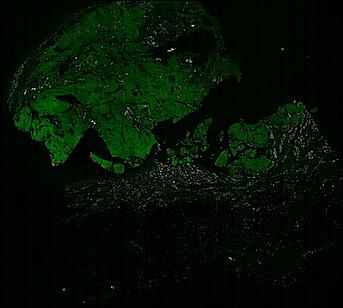
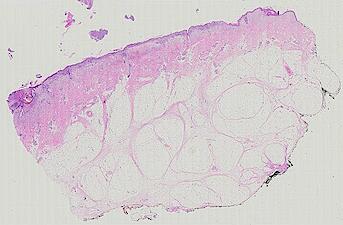
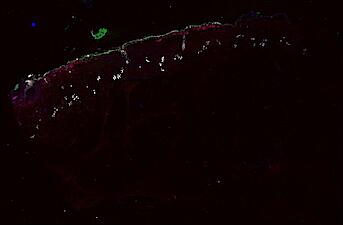
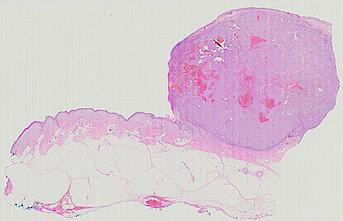
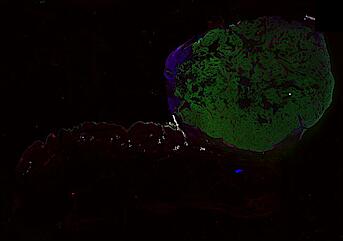
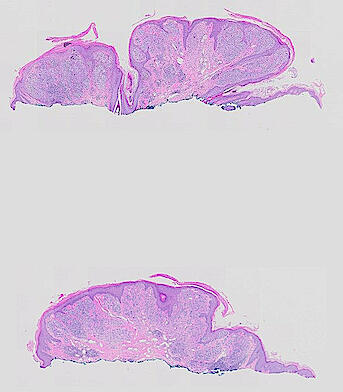
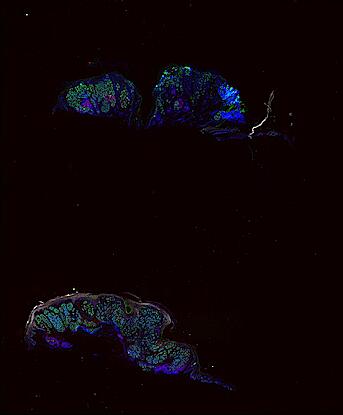
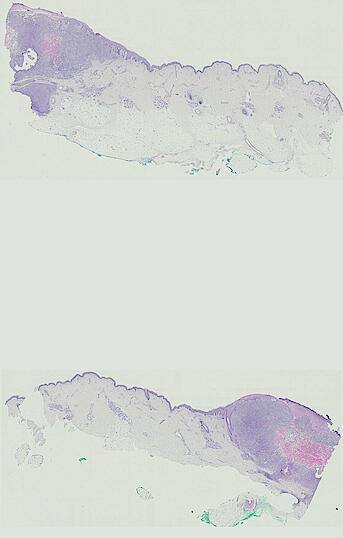
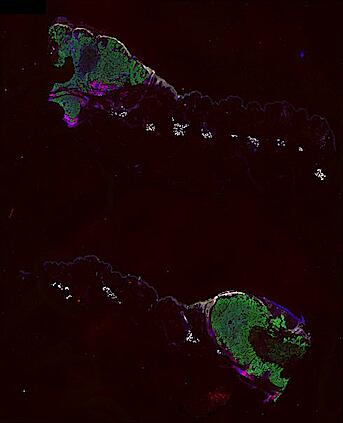
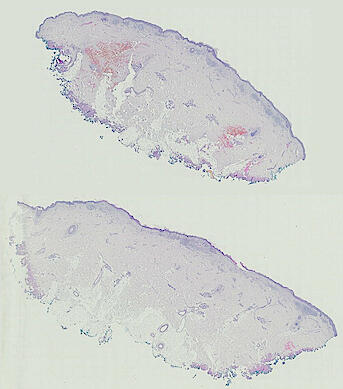
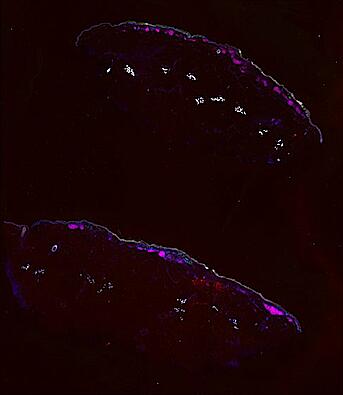
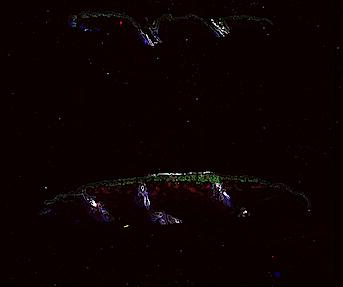
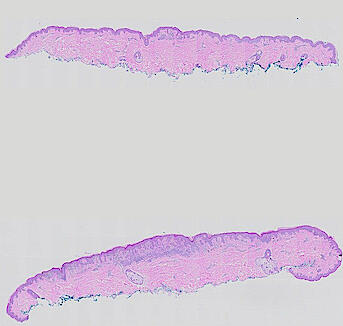
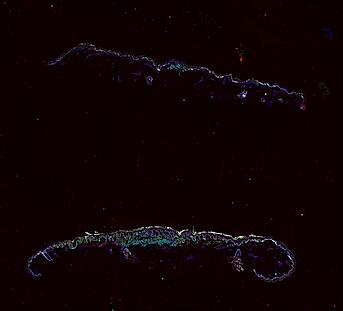
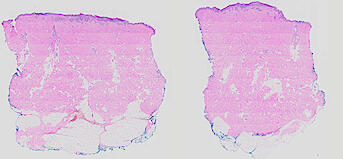
The spatial landscape of progression and immunoediting in primary melanoma at single cell resolution
Nirmal AJ, Maliga Z, Vallius T, Quattrochi B, Chen AC, Jacobson CA, Pelletier RJ, ... Lian CG, Murphy GF, Santagata S, Sorger PK
Cancer Discovery. 2022 Jun; 12(6): 1518-1541. PMID: 35404441
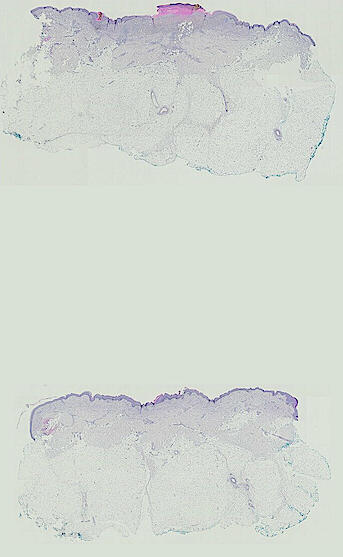
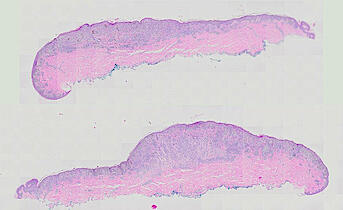
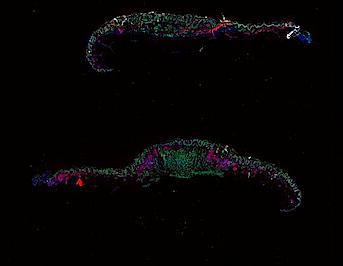
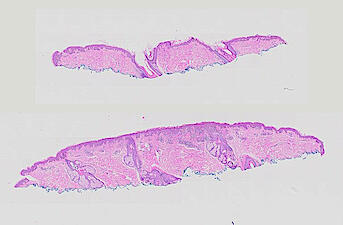
Cutaneous melanoma is a highly immunogenic malignancy, surgically curable at early stages, but life- threatening when metastatic. Here we integrate high-plex imaging, 3D high-resolution microscopy, and spatially-resolved micro-region transcriptomics to study immune evasion and immunoediting in primary melanoma. We find that recurrent cellular neighborhoods involving tumor, immune, and stromal cells change significantly along a progression axis involving precursor states, melanoma in situ, and invasive tumor. Hallmarks of immunosuppression are already detectable in precursor regions. When tumors become locally invasive, a consolidated and spatially restricted suppressive environment forms along the tumor-stromal boundary. This environment is established by cytokine gradients that promote expression of MHC-II and IDO1, and by PD1-PDL1 mediated cell contacts involving macrophages, dendritic cells, and T cells. A few millimeters away, cytotoxic T cells synapse with melanoma cells in fields of tumor regression. Thus, invasion and immunoediting can co-exist within a few millimeters of each other in a single specimen.
Publication bioRxiv Melanoma atlas Access Primary DataNarrated Minerva Stories
Narrated stories use multi-step narrations and annotations to walk a viewer through key features of the data. Narrated stories distill the multidisciplinary knowledge encompassed by each dataset into a single product that grounds the scientific analyses in the underlying data and metadata. Click the Minerva story icon for an interactive view of the full-resolution images.
Automated Minerva Stories
Automated stories provide basic image viewing with automatic rendering settings, enabling rapid, lightweight sharing of highly multiplexed images, without download. Click the Minerva story icon for an interactive view of the full-resolution images.
Primary Data Access
About the data files
The primary data represents minimally processed (Level 2) image data from either 2D whole slide or 3D optically-sectioned imaging relevant to HMS HTAN Center Melanoma Atlas 1. Whole slide images are segmented, quantified, and subjected to additional quality control to generate final data. Whole slide scans are saved as OME-TIFF tiled pyramid images whereas 3D datasets are three-dimensional TIFF files. Spatial feature tables are zipped CSV files.
Data were collected using cyclic immunofluorescence (CyCIF), as described in https://dx.doi.org/10.17504/protocols.io.bjiukkew, or brightfield imaging of hematoxylin and eosin (H&E) stained slides. Each file corresponds to a multiplexed image mosaic for FFPE tissue sections 5 microns thick with sample diameter extending up to ~2.9 cm. Each whole slide image is assembled from a series of successive image tiles stitched together (832 x 732 µm tiles; up to 990 tiles/image) and flat-field corrected for illumination using MCMICRO software to generate ~0.2 to ~1.5 gigapixel images. Tiles were collected for each CyCIF round (up to 15 in total depending of the antibody panel used) and these are combined in the mosaic image to generate a composite with 32 to 60 channels. Each 3D image (110 x 110 x 16 µm) requires 3D image registration to assemble all 7 CyCIF rounds.
Whole slide CyCIF images were collected on a RareCyte Inc. CyteFinder slide scanning fluorescence microscope using a 20x/0.75 NA objective and sampled at 650 nm/pixel. 3D high resolution CyCIF images were acquired on a GE Deltavision Elite equipped with a 60x/1.42 NA oil immersion objective lens and sampled at 108 nm/pixel in X & Y, and 200 nm steps in Z axis. H&E images were collected on an Olympus VS120 microscope using a 20x/0.75 NA objective and sampled at 350 nm/pixel.
The files MEL01-1-* to MEL01-3-* derive from a 62-year old male (MEL1), who had a stage IIC primary melanoma with NF1 (c.1008G>A and c.4006C>T) mutation. The tumor had invaded into reticular dermis and was surgically removed.
The files MEL02-1-* to MEL13-2-* derive from 12 additional patients from the Brigham and Women’s Hospital. Additional information on antibodies and specimen are available at https://labsyspharm.github.io/HTA-MELATLAS-1/.
Image files ending in -0-ROI* are H&E images. All others are CyCIF images.
Download the primary data
Download the primary data
The full dataset is available through the Human Tumor Atlas Network. The Laboratory of Systems Pharmacology also makes the data available through Amazon Web Services S3. The images and metadata will be available in the bucket at the following location:
s3://hta-melatlas-1/data/
Follow our instructions on Zenodo to access this dataset (DOI: 10.5281/zenodo.10223574).
The following table contains summary biospecimen and file metadata.
Whole-slide images (OME-TIFF) and spatial feature tables (CSV)
| FIELD1 | Level 2 WSI (OME-TIFF) | File size (WSI) | Spatial feature tables (CSV) | File size (feature table) |
|---|---|---|---|---|
| MEL01-1-0-ROI1 | MEL01-1-0-HE-ROI1.ome.tif | 15 GB | ||
| MEL01-1-1 | MEL01-1-1.ome.tif | 195 GB | ||
| MEL01-1-3 | MEL01-1-3.ome.tif | 98 GB | MEL01-1-3-features.zip | 335 MB |
| MEL01-1-4 | MEL01-1-4.ome.tif | 66 GB | MEL01-1-4-features.zip | 200 MB |
| MEL01-1-5 | MEL01-1-5.ome.tif | 129 GB | MEL01-1-5-features.zip | 280 MB |
| MEL01-1-6 | MEL01-1-6.ome.tif | 174 GB | MEL01-1-6-features.zip | 290 MB |
| MEL01-2-0-ROI1 | MEL01-2-0-HE-ROI1.ome.tif | 13 GB | ||
| MEL01-2-1 | MEL01-2-1.ome.tif | 163 GB | MEL01-2-1-features.zip | 82 MB |
| MEL01-3-0-ROI1 | MEL01-3-0-HE-ROI1.ome.tif | 17 GB | ||
| MEL01-3-1 | MEL01-3-1.ome.tif | 230 GB | MEL01-3-1-features.zip | 280 MB |
| MEL02-1-0-ROI1 | MEL02-1-0-HE-ROI1.ome.tif | 0.5 GB | ||
| MEL02-1-0-ROI2 | MEL02-1-0-HE-ROI2.ome.tif | 0.4 GB | ||
| MEL02-1-1 | MEL02-1-1.ome.tif | 19 GB | MEL02-1-1-features.zip | 16 MB |
| MEL03-1-0-ROI1 | MEL03-1-0-HE-ROI1.ome.tif | 6 GB | ||
| MEL03-1-0-ROI2 | MEL03-1-0-HE-ROI2.ome.tif | 5 GB | ||
| MEL03-1-1 | MEL03-1-1.ome.tif | 189 GB | MEL03-1-1-features.zip | 160 MB |
| MEL04-1-0-ROI1 | MEL04-1-0-HE-ROI1.ome.tif | 5 GB | ||
| MEL04-1-0-ROI2 | MEL04-1-0-HE-ROI2.ome.tif | 4 GB | ||
| MEL04-1-1 | MEL04-1-1.ome.tif | 179 GB | MEL04-1-1-features.zip | 61 MB |
| MEL05-1-0-ROI1 | MEL05-1-0-HE-ROI1.ome.tif | 0.4 GB | ||
| MEL05-1-0-ROI2 | MEL05-1-0-HE-ROI2.ome.tif | 0.6 GB | ||
| MEL05-1-1 | MEL05-1-1.ome.tif | 34 GB | MEL05-1-1-features.zip | 17 MB |
| MEL06-1-0-ROI1 | MEL06-1-0-HE-ROI1.ome.tif | 17 GB | ||
| MEL06-1-1 | MEL06-1-1.ome.tif | 220 GB | MEL06-1-1-features.zip | 299 MB |
| MEL07-1-0-ROI1 | MEL07-1-0-HE-ROI1.ome.tif | 4 GB | ||
| MEL07-1-0-ROI2 | MEL07-1-0-HE-ROI2.ome.tif | 3 GB | ||
| MEL07-1-1 | MEL07-1-1.ome.tif | 122 GB | MEL07-1-1-features.zip | 41 MB |
| MEL08-1-0-ROI1 | MEL08-1-0-HE-ROI1.ome.tif | 1 GB | ||
| MEL08-1-0-ROI2 | MEL08-1-0-HE-ROI2.ome.tif | 0.9 GB | ||
| MEL08-1-1 | MEL08-1-1.ome.tif | 40 GB | MEL08-1-1-features.zip | 29 MB |
| MEL09-1-0-ROI1 | MEL09-1-0-HE-ROI1.ome.tif | 0.8 GB | ||
| MEL09-1-0-ROI2 | MEL09-1-0-HE-ROI2.ome.tif | 0.6 GB | ||
| MEL09-1-1 | MEL09-1-1.ome.tif | 32 GB | MEL09-1-1-features.zip | 6 MB |
| MEL10-1-0-ROI1 | MEL10-1-0-HE-ROI1.ome.tif | 0.5 GB | ||
| MEL10-1-0-ROI2 | MEL10-1-0-HE-ROI2.ome.tif | 0.6 GB | ||
| MEL10-1-1 | MEL10-1-1.ome.tif | 34 GB | MEL10-1-1-features.zip | 12 MB |
| MEL11-1-0-ROI1 | MEL11-1-0-HE-ROI1.ome.tif | 2 GB | ||
| MEL11-1-0-ROI2 | MEL11-1-0-HE-ROI2.ome.tif | 2 GB | ||
| MEL11-1-1 | MEL11-1-1.ome.tif | 79 GB | MEL11-1-1-features.zip | 21 MB |
| MEL12-1-0-ROI1 | MEL12-1-0-HE-ROI1.ome.tif | 2 GB | ||
| MEL12-1-0-ROI2 | MEL12-1-0-HE-ROI2.ome.tif | 1 GB | ||
| MEL12-1-1 | MEL12-1-1.ome.tif | 56 GB | MEL12-1-1-features.zip | 14 MB |
| MEL13-1-0-ROI1 | MEL13-1-0-HE-ROI1.ome.tif | 4 GB | ||
| MEL13-1-0-ROI2 | MEL13-1-0-HE-ROI2.ome.tif | 3 GB | ||
| MEL13-1-1 | MEL13-1-1.ome.tif | 129 GB | MEL13-1-1-features.zip | 46 MB |
| MEL13-2-0-ROI1 | MEL13-2-0-HE-ROI1.ome.tif | 4 GB | ||
| MEL13-2-0-ROI2 | MEL13-2-0-HE-ROI2.ome.tif | 3 GB | ||
| MEL13-2-1 | MEL13-2-1.ome.tif | 121 GB | MEL13-2-1-features.zip | 36 MB |
3D high-resolution images
| FIELD1 | 3D image file | File size |
|---|---|---|
| MEL01-1-4-3D-1 | MEL01-1-4-3D-1.ome.tif | 5 GB |
| MEL01-1-4-3D-10 | MEL01-1-4-3D-10.ome.tif | 5 GB |
| MEL01-1-4-3D-11 | MEL01-1-4-3D-11.ome.tif | 5 GB |
| MEL01-1-4-3D-12 | MEL01-1-4-3D-12.ome.tif | 4 GB |
| MEL01-1-4-3D-13 | MEL01-1-4-3D-13.ome.tif | 5 GB |
| MEL01-1-4-3D-14 | MEL01-1-4-3D-14.ome.tif | 5 GB |
| MEL01-1-4-3D-15 | MEL01-1-4-3D-15.ome.tif | 5 GB |
| MEL01-1-4-3D-16 | MEL01-1-4-3D-16.ome.tif | 5 GB |
| MEL01-1-4-3D-17 | MEL01-1-4-3D-17.ome.tif | 5 GB |
| MEL01-1-4-3D-18 | MEL01-1-4-3D-18.ome.tif | 5 GB |
| MEL01-1-4-3D-19 | MEL01-1-4-3D-19.ome.tif | 8 GB |
| MEL01-1-4-3D-2 | MEL01-1-4-3D-2.ome.tif | 5 GB |
| MEL01-1-4-3D-20 | MEL01-1-4-3D-20.ome.tif | 5 GB |
| MEL01-1-4-3D-21 | MEL01-1-4-3D-21.ome.tif | 5 GB |
| MEL01-1-4-3D-22 | MEL01-1-4-3D-22.ome.tif | 5 GB |
| MEL01-1-4-3D-23 | MEL01-1-4-3D-23.ome.tif | 4 GB |
| MEL01-1-4-3D-24 | MEL01-1-4-3D-24.ome.tif | 5 GB |
| MEL01-1-4-3D-25 | MEL01-1-4-3D-25.ome.tif | 5 GB |
| MEL01-1-4-3D-26 | MEL01-1-4-3D-26.ome.tif | 5 GB |
| MEL01-1-4-3D-27 | MEL01-1-4-3D-27.ome.tif | 5 GB |
| MEL01-1-4-3D-28 | MEL01-1-4-3D-28.ome.tif | 4 GB |
| MEL01-1-4-3D-29 | MEL01-1-4-3D-29.ome.tif | 4 GB |
| MEL01-1-4-3D-3 | MEL01-1-4-3D-3.ome.tif | 5 GB |
| MEL01-1-4-3D-30 | MEL01-1-4-3D-30.ome.tif | 5 GB |
| MEL01-1-4-3D-31 | MEL01-1-4-3D-31.ome.tif | 4 GB |
| MEL01-1-4-3D-32 | MEL01-1-4-3D-32.ome.tif | 5 GB |
| MEL01-1-4-3D-33 | MEL01-1-4-3D-33.ome.tif | 4 GB |
| MEL01-1-4-3D-34 | MEL01-1-4-3D-34.ome.tif | 4 GB |
| MEL01-1-4-3D-35 | MEL01-1-4-3D-35.ome.tif | 4 GB |
| MEL01-1-4-3D-36 | MEL01-1-4-3D-36.ome.tif | 4 GB |
| MEL01-1-4-3D-37 | MEL01-1-4-3D-37.ome.tif | 4 GB |
| MEL01-1-4-3D-38 | MEL01-1-4-3D-38.ome.tif | 4 GB |
| MEL01-1-4-3D-39 | MEL01-1-4-3D-39.ome.tif | 4 GB |
| MEL01-1-4-3D-4 | MEL01-1-4-3D-4.ome.tif | 5 GB |
| MEL01-1-4-3D-40 | MEL01-1-4-3D-40.ome.tif | 6 GB |
| MEL01-1-4-3D-41 | MEL01-1-4-3D-41.ome.tif | 4 GB |
| MEL01-1-4-3D-42 | MEL01-1-4-3D-42.ome.tif | 4 GB |
| MEL01-1-4-3D-5 | MEL01-1-4-3D-5.ome.tif | 5 GB |
| MEL01-1-4-3D-6 | MEL01-1-4-3D-6.ome.tif | 5 GB |
| MEL01-1-4-3D-7 | MEL01-1-4-3D-7.ome.tif | 5 GB |
| MEL01-1-4-3D-8 | MEL01-1-4-3D-8.ome.tif | 5 GB |
| MEL01-1-4-3D-9 | MEL01-1-4-3D-9.ome.tif | 4 GB |
 Click to enlarge.
Click to enlarge.