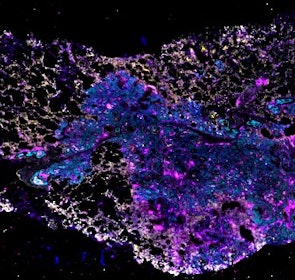
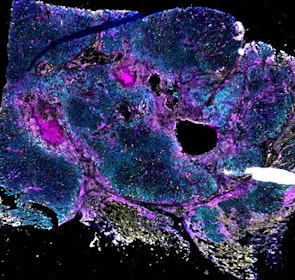
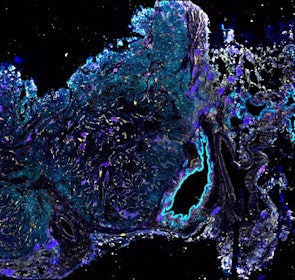
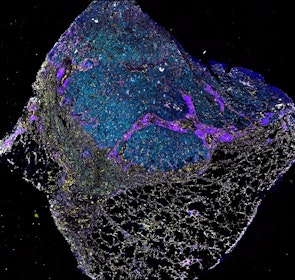
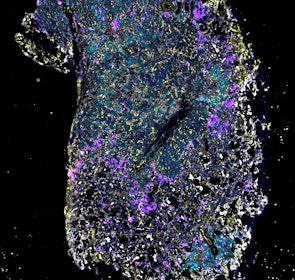
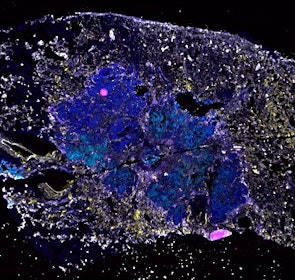
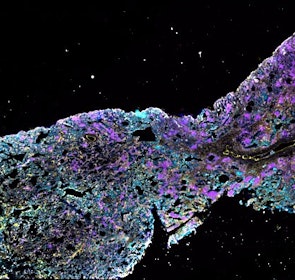
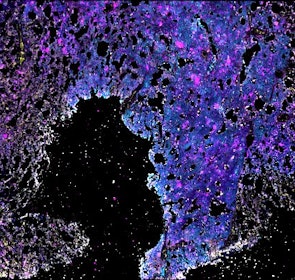
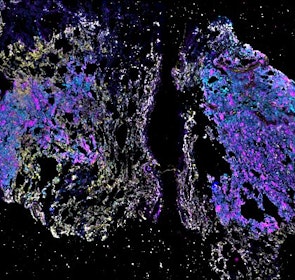
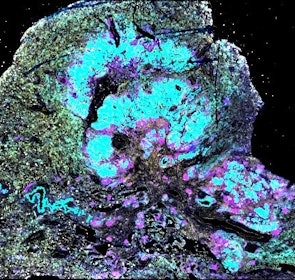
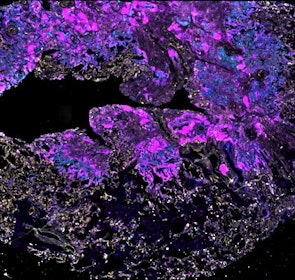
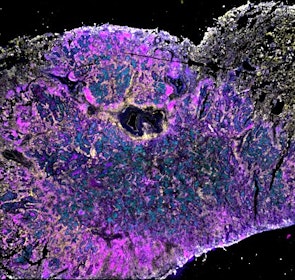
Spatial intra-tumor heterogeneity is associated with survival of lung adenocarcinoma patients
Wu HJ, Temko D, Maliga M, Moreira A, Sei E, Conterno Minussi D, Dean J, Lee C, Xu Q, Hochart G, Jacobson C, Yapp C, Schapiro D, Sorger P, Seeley EH, Navin N, Downey RJ, and Michor F
Cell Genomics. 2022 Aug; 2(8): 100165. PMID: 35404441
Intratumor heterogeneity (ITH) of human tumors is important for tumor progression, treatment response, and drug resistance. However, the spatial distribution of ITH remains incompletely understood. Here, we present spatial analysis of ITH in lung adenocarcinomas from 147 patients using multi-region mass spectrometry of >5000 regions, single cell copy number sequencing of ~2000 single cells, and cyclic immunofluorescence of >10 million cells. We identified two distinct spatial patterns among tumors, termed clustered and random geographic diversification (GD). These patterns were observed in the same samples using both proteomic and genomic data. The random proteomic GD pattern, which is characterized by decreased cell adhesion and lower levels of tumor-interacting endothelial cells, was significantly associated with increased risk of recurrence or death in two independent patient cohorts. Our study presents comprehensive spatial mapping of ITH in lung adenocarcinoma and provides insights into the mechanisms and clinical consequences of geographic diversification of intratumor heterogeneity.
Publication Access Primary DataAutomated Minerva Stories
Automated stories provide basic image viewing with automatic rendering settings, enabling rapid, lightweight sharing of highly multiplexed images, without download. Click the Minerva story icon for an interactive view of the full-resolution images.
Data Access
About the Data Files
This dataset can be used to reproduce the analysis of 12 lung tumors, as described (Wu HJ, Temko D, Maliga Z, et. al. Cell Genomics, 2022), and to develop new computational tools for processing and analysis of tumors stained for standard histopathology (hematoxylin and eosin) or highly multiplexed (20+) fluorescent tissue imaging.
The dataset includes raw brightfield and immunofluorescence images of surgical resections of lung adenocarcinoma from twelve patients. The stitched image pyramids for highly multiplexed fluorescence image data, segmentation masks and single-cell intensity files for each sample generated by MCMICRO is included.
The position of histologically defined regions (20-60 per tumor) in the H&E stained sections and the corresponding region in the adjacent section stained by multiplexed fluorescence microscopy (CyCIF) are indicated.
Download the primary data
Download the primary data
File Types
Each folder corresponds to a patient sample (N). The following files are available for each patient and are located on Synapse (synID syn32529019) or on Amazon Web Services (AWS).
Free account registration is required to download files from Synapse. Files available through AWS S3 are available in the bucket at the following location: s3://wu-temko-maliga-2022
| File Type | Description | Location |
|---|---|---|
| N.ome.tif | Stitched multiplex CyCIF image pyramid in ome.tif format | AWS |
| N_HE.vsi | Hematoxylin and Eosin stained image of adjacent FFPE tissue section in .vsi format | AWS |
| _N_HE_/ | folder: raw image data accompanying .vsi file | AWS |
| markers.csv | list of all markers in ome.tif image | Synapse |
| N.csv | single-cell feature table, including intensity data for all channels | Synapse |
| N_ROI.csv | X and Y coordinates for histologically annotated regions in CyCIF and H&E images | Synapse |
| raw/ | folder of raw IF image data in .rcpnl format | AWS |
| segmentation/ | folder of segmentation maps for tissue image in .tif format | AWS |
N.ome.tif
| Patient | File Name | Location | File size |
|---|---|---|---|
| P132115 | P132115.ome.tif | AWS | 90.6 GB |
| P132630 | P132630.ome.tif | AWS | 99.9 GB |
| P132666 | P132666.ome.tif | AWS | 123.1 GB |
| P133537 | P133537.ome.tif | AWS | 87.8 GB |
| P136690 | P136690.ome.tif | AWS | 56.5 GB |
| P137591 | P137591.ome.tif | AWS | 91.8 GB |
| P137753 | P137753.ome.tif | AWS | 144.4 GB |
| P137757 | P137757.ome.tif | AWS | 100.8 GB |
| P137905 | P137905.ome.tif | AWS | 132.4 GB |
| P137941 | P137941.ome.tif | AWS | 121.0 GB |
| P137974 | P137974.ome.tif | AWS | 134.6 GB |
| P138007 | P138007.ome.tif | AWS | 128.6 GB |
N_HE.vsi
| Patient | File Name | Location | File size |
|---|---|---|---|
| P132115 | P132115_HE.vsi | AWS | 531.2 KB |
| P132630 | P132630_HE.vsi | AWS | 845.8 KB |
| P132666 | P132666_HE.vsi | AWS | 592.7 KB |
| P133537 | P133537_HE.vsi | AWS | 948.9 KB |
| P136690 | P136690_HE.vsi | AWS | 866.2 KB |
| P137591 | P137591_HE.vsi | AWS | 625.6 KB |
| P137753 | P137753_HE.vsi | AWS | 540.9 KB |
| P137757 | P137757_HE.vsi | AWS | 742.2 KB |
| P137905 | P137905_HE.vsi | AWS | 685.5 KB |
| P137941 | P137941_HE.vsi | AWS | 683.5 KB |
| P137974 | P137974_HE.vsi | AWS | 636.0 KB |
| P138007 | P138007_HE.vsi | AWS | 678.0 KB |
_N_HE_/
| Patient | File Name | Location | File size |
|---|---|---|---|
| P132115 | frame_t.ets | AWS | 928.7 MB |
| P132630 | frame_t.ets | AWS | 1.3 GB |
| P132666 | frame_t.ets | AWS | 1.5 GB |
| P133537 | frame_t.ets | AWS | 980.1 MB |
| P136690 | frame_t.ets | AWS | 792.1 MB |
| P137591 | frame_t.ets | AWS | 1.1 GB |
| P137753 | frame_t.ets | AWS | 1.5 GB |
| P137757 | frame_t.ets | AWS | 1.6 GB |
| P137905 | frame_t.ets | AWS | 1.4 GB |
| P137941 | frame_t.ets | AWS | 1.8 GB |
| P137974 | frame_t.ets | AWS | 1.6 GB |
| P138007 | frame_t.ets | AWS | 2.0 GB |
markers.csv
| Patient | File Name | Synapse ID | File size |
|---|---|---|---|
| P132115 | markers.csv | syn32563757 | 530 bytes |
| P132630 | markers.csv | syn32563890 | 579 bytes |
| P132666 | markers.csv | syn32564039 | 530 bytes |
| P133537 | markers.csv | syn32564369 | 579 bytes |
| P136690 | markers.csv | syn32564613 | 579 bytes |
| P137591 | markers.csv | syn32564701 | 530 bytes |
| P137753 | markers.csv | syn32565266 | 530 bytes |
| P137757 | markers.csv | syn32565460 | 437 bytes |
| P137905 | markers.csv | syn32565554 | 530 bytes |
| P137941 | markers.csv | syn32565759 | 530 bytes |
| P137974 | markers.csv | syn32566194 | 623 bytes |
| P138007 | markers.csv | syn32566617 | 623 bytes |
N.csv
| Patient | File Name | Synapse ID | File size |
|---|---|---|---|
| P132115 | P132115.csv | syn32563869 | 624.7 MB |
| P132630 | P132630.csv | syn32564017 | 960.1 MB |
| P132666 | P132666.csv | syn32564305 | 1.107 GB |
| P133537 | P133537.csv | syn32564460 | 622.8 MB |
| P136690 | P136690.csv | syn32564683 | 503.3 MB |
| P137591 | P137591.csv | syn32564818 | 574.3 MB |
| P137753 | P137753.csv | syn32565424 | 1.064 GB |
| P137905 | P137905.csv | syn32565737 | 896.9 MB |
| P137757 | P137757.csv | syn32565509 | 529.4 MB |
| P137941 | P137941.csv | syn32566154 | 1.338 GB |
| P137974 | P137974.csv | syn32566607 | 1.073 GB |
| P138007 | P138007.csv | syn32566788 | 1.339 GB |
N_ROI.csv
| Patient | File Name | Synapse ID | File size |
|---|---|---|---|
| P132115 | P132115_ROI.csv | syn32563870 | 2.2 KB |
| P132630 | P132630_ROI.csv | syn32564019 | 4.5 KB |
| P132666 | P132666_ROI.csv | syn32564307 | 2.6 KB |
| P133537 | P133537_ROI.csv | syn32564462 | 3.2 KB |
| P136690 | P136690_ROI.csv | syn32564685 | 3.7 KB |
| P137591 | P137591_ROI.csv | syn32564821 | 3.5 KB |
| P137753 | P137753_ROI.csv | syn32565425 | 2.9 KB |
| P137757 | P137757_ROI.csv | syn32565511 | 1.5 KB |
| P137905 | P137905_ROI.csv | syn32565738 | 2.3 KB |
| P137941 | P137941_ROI.csv | syn32566155 | 1.5 KB |
| P137974 | P137974_ROI.csv | syn32566609 | 2.5 KB |
| P138007 | P138007_ROI.csv | syn32566789 | 2.1 KB |
raw/
| Patient | Number of Files | Folder size | Location |
|---|---|---|---|
| P132115 | 13 | 78.9 GB | AWS |
| P132630 | 13 | 86.8 GB | AWS |
| P132666 | 13 | 109.3 GB | AWS |
| P133537 | 13 | 77.1 GB | AWS |
| P136690 | 13 | 48.2 GB | AWS |
| P137591 | 13 | 81.5 GB | AWS |
| P137753 | 13 | 125.3 GB | AWS |
| P137757 | 9 | 75.6 GB | AWS |
| P137905 | 13 | 115.7 GB | AWS |
| P137941 | 13 | 106.1 GB | AWS |
| P137974 | 13 | 118.9 GB | AWS |
| P138007 | 13 | 112.5 GB | AWS |
segmentation/
| Patient | Number of Files | Folder size | Location |
|---|---|---|---|
| P132115 | 12 | 35.3 GB | AWS |
| P132630 | 12 | 38.8 GB | AWS |
| P132666 | 12 | 48.9 GB | AWS |
| P133537 | 12 | 34.5 GB | AWS |
| P136690 | 12 | 21.6 GB | AWS |
| P137591 | 12 | 36.5 GB | AWS |
| P137753 | 12 | 56.1 GB | AWS |
| P137757 | 12 | 48.9 GB | AWS |
| P137905 | 12 | 51.8 GB | AWS |
| P137941 | 12 | 47.5 GB | AWS |
| P137974 | 12 | 53.2 GB | AWS |
| P138007 | 12 | 50.3 GB | AWS |