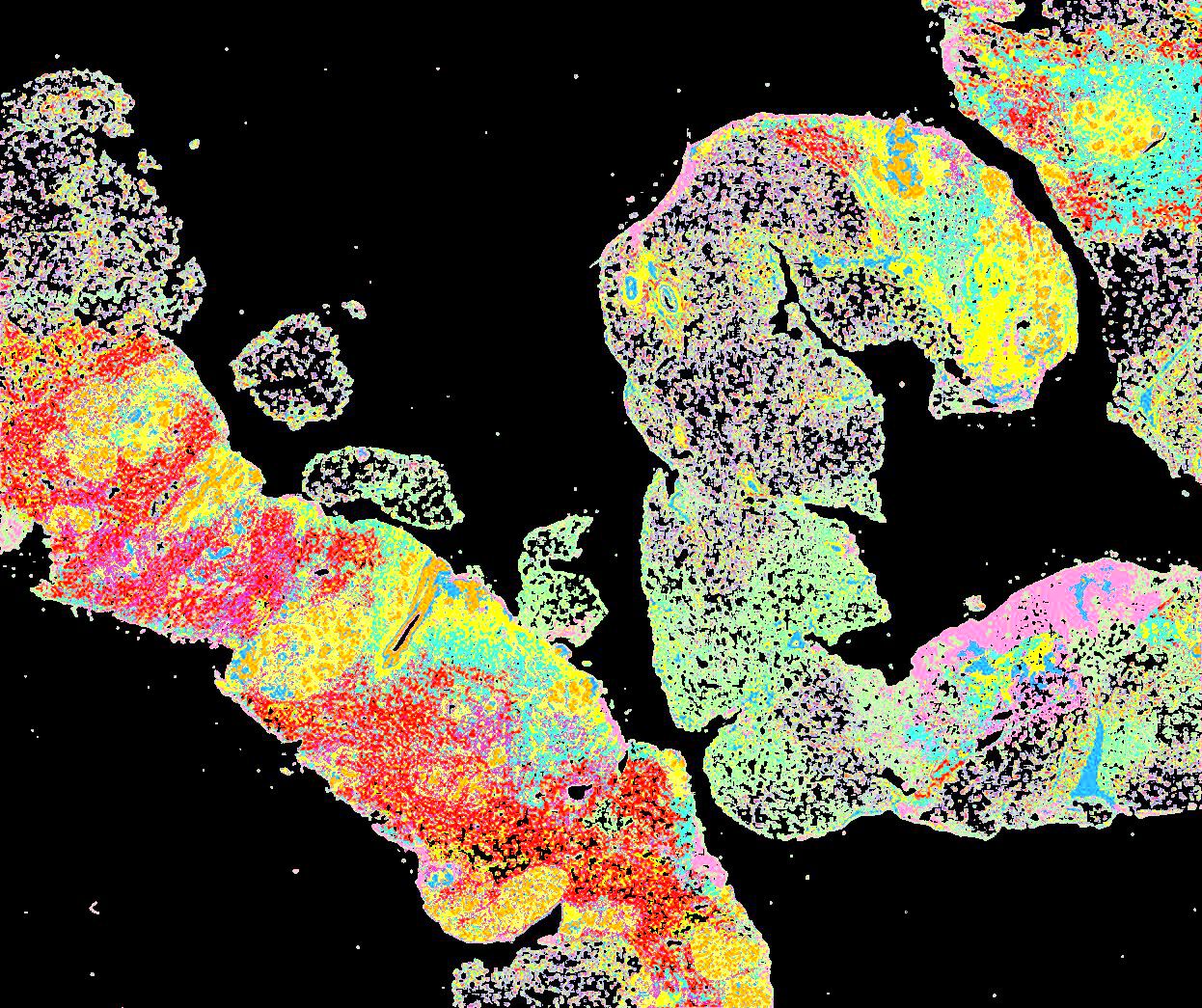
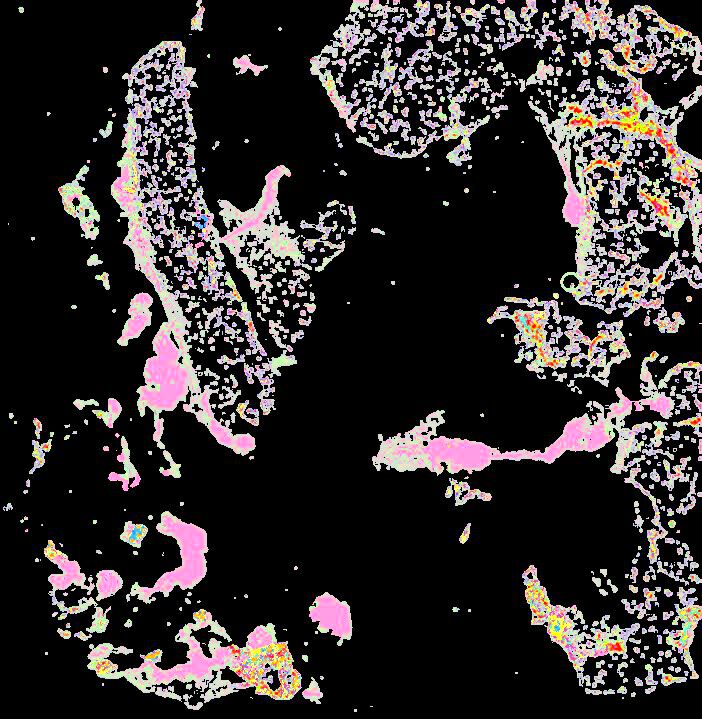
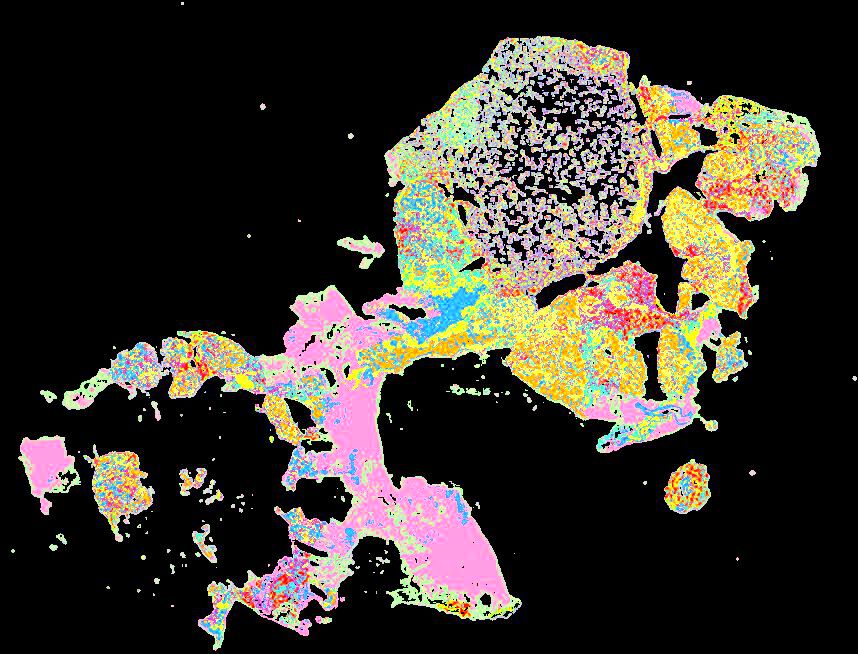
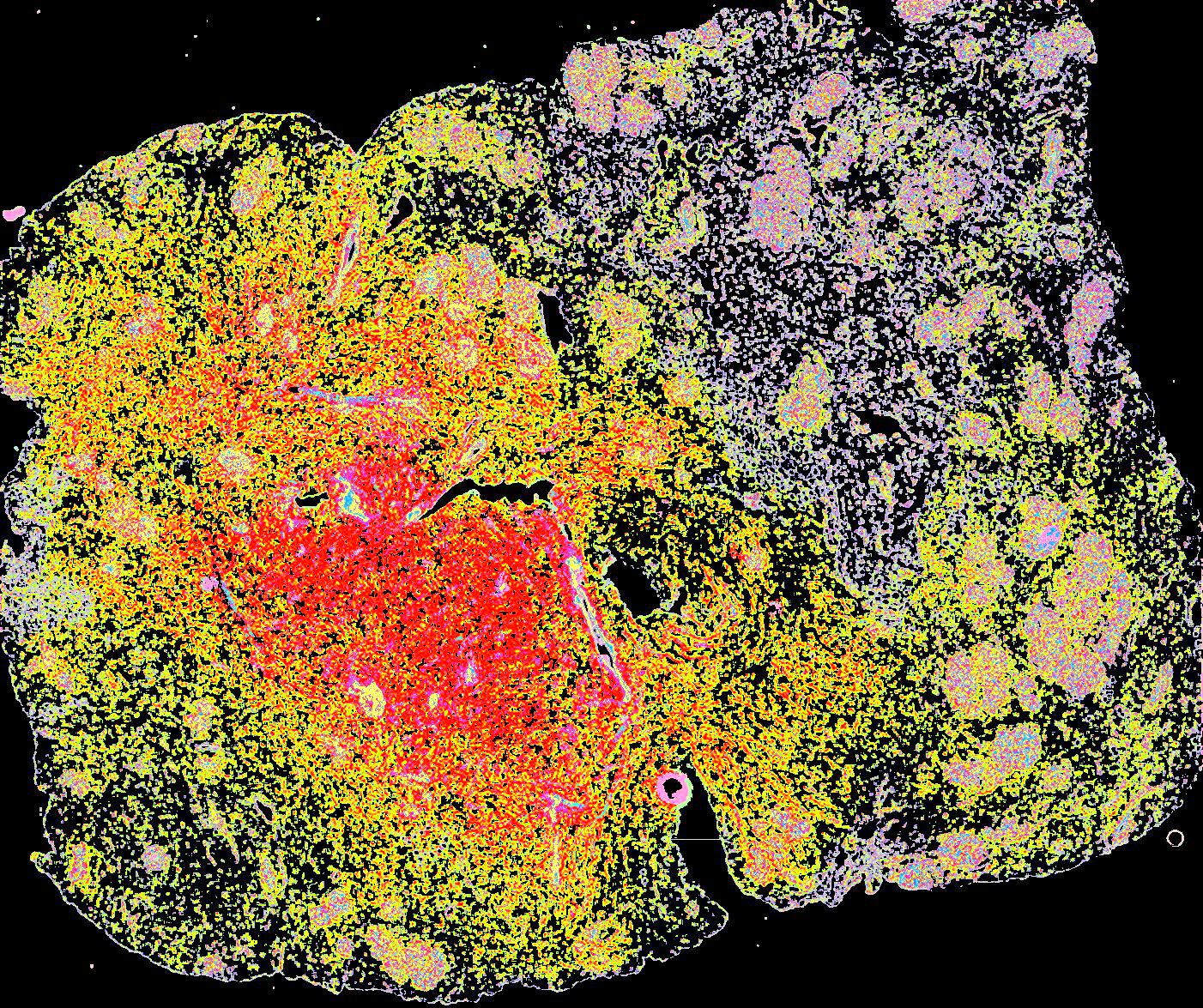
A human breast atlas integrating single-cell proteomics and transcriptomics
Gray GK, Li CM, Rosenbluth JM, Selfors LM, Girnius N, Lin JR, Schackmann RCJ, Goh WL, Moore K, Shapiro HK, Mei S, DAndrea K, Nathanson KL, Sorger PK, Santagata S, Regev A, Garber JE, Dillon DA, and Brugge JS
Developmental Cell. 2022 June 6; 57(11): 1400-1420.e7. DOI: 10.1016/j.devcel.2022.05.003
The breast is a dynamic organ whose response to physiological and pathophysiological conditions alters its disease susceptibility, yet the specific effects of these clinical variables on cell state remain poorly annotated. We present a unified, high-resolution breast atlas by integrating single-cell RNA-seq, mass cytometry, and cyclic immunofluorescence, encompassing a myriad of states. We define cell subtypes within the alveolar, hormone-sensing, and basal epithelial lineages, delineating associations of several subtypes with cancer risk factors, including age, parity, and BRCA2 germline mutation. Of particular interest is a subset of alveolar cells termed basal-luminal (BL) cells, which exhibit poor transcriptional lineage fidelity, accumulate with age, and carry a gene signature associated with basal-like breast cancer. We further utilize a medium-depletion approach to identify molecular factors regulating cell-subtype proportion in organoids. Together, these data are a rich resource to elucidate diverse mammary cell states.
Publication Sequencing CyTOF Gray BRCA AtlasKey Findings
-
Multimodal single-cell analyses identify breast epithelial and stromal subtypes
-
Spatially distinct epithelial subsets are linked with age, parity, and BRCA2 status
-
Alveolar cells with poor transcriptional lineage fidelity accumulate with age
-
Subtypes of the three major epithelial lineages are maintained in organoid cultures
Curated Minerva Stories
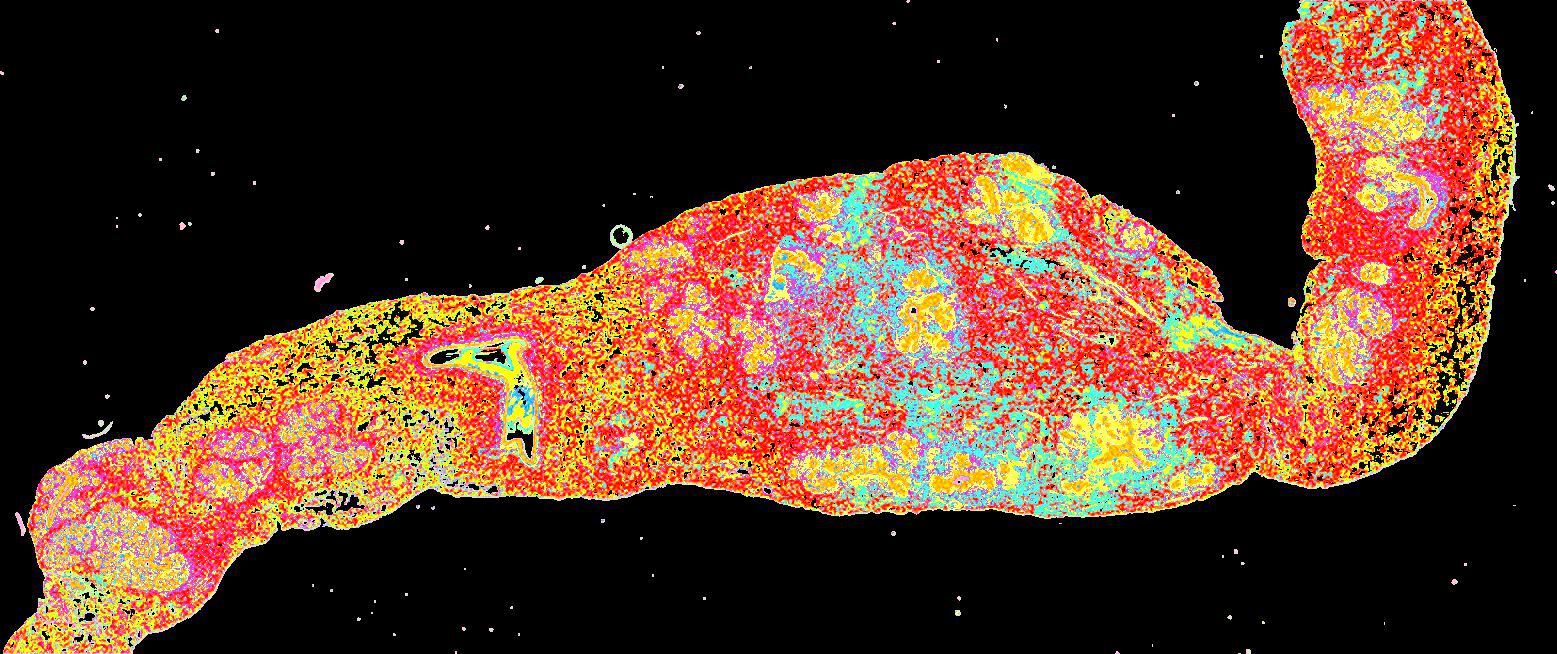
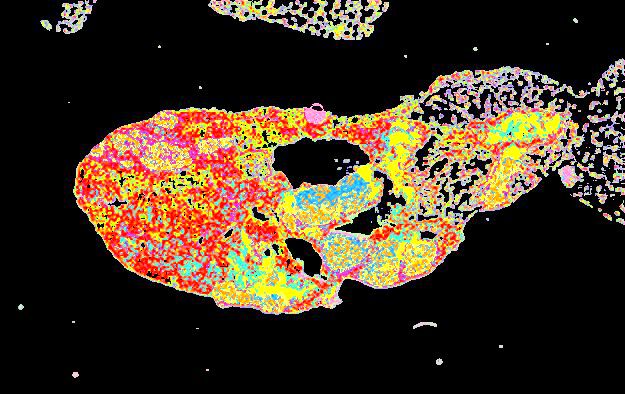
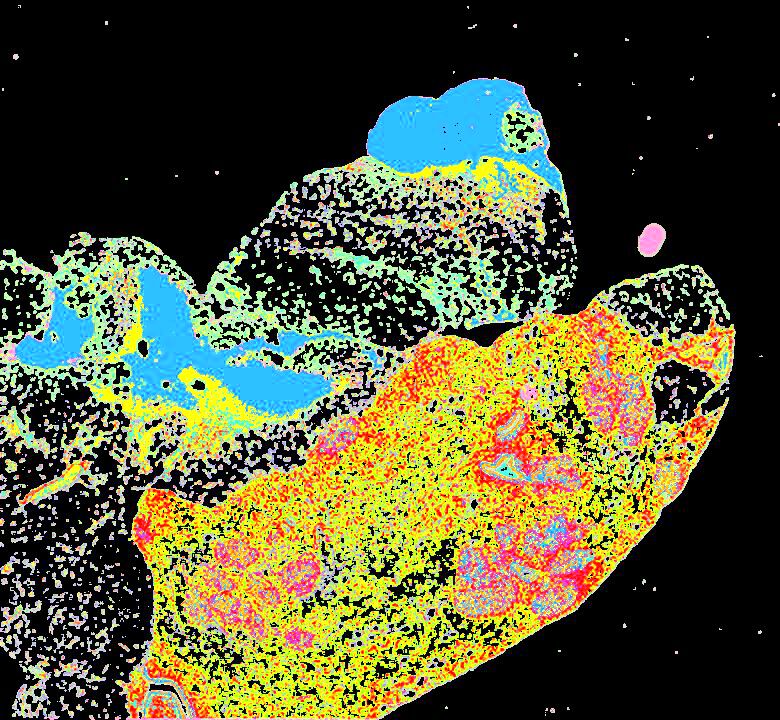
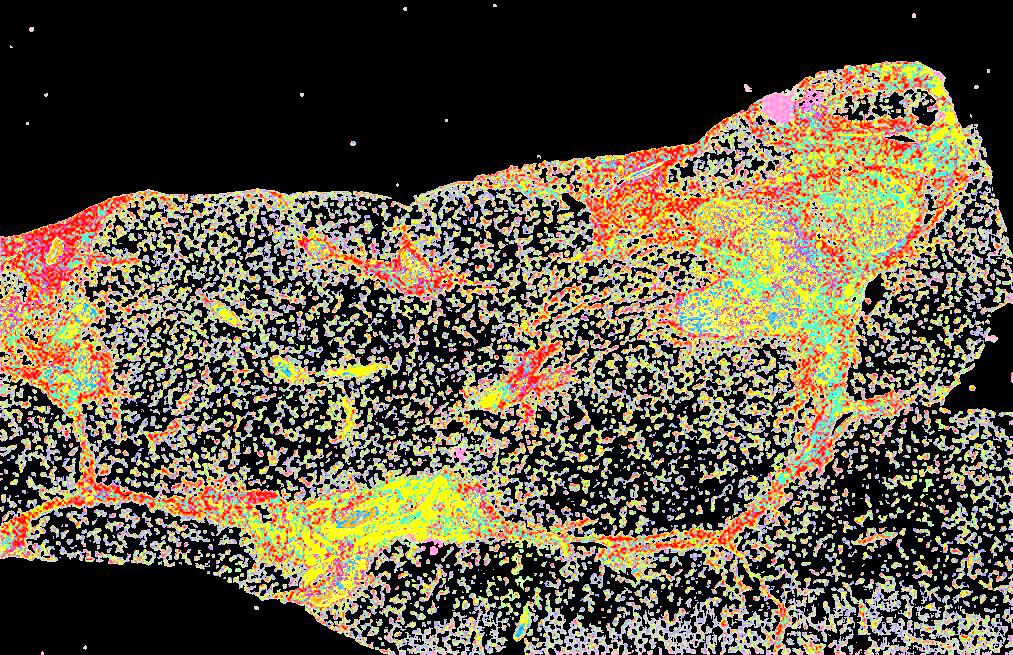
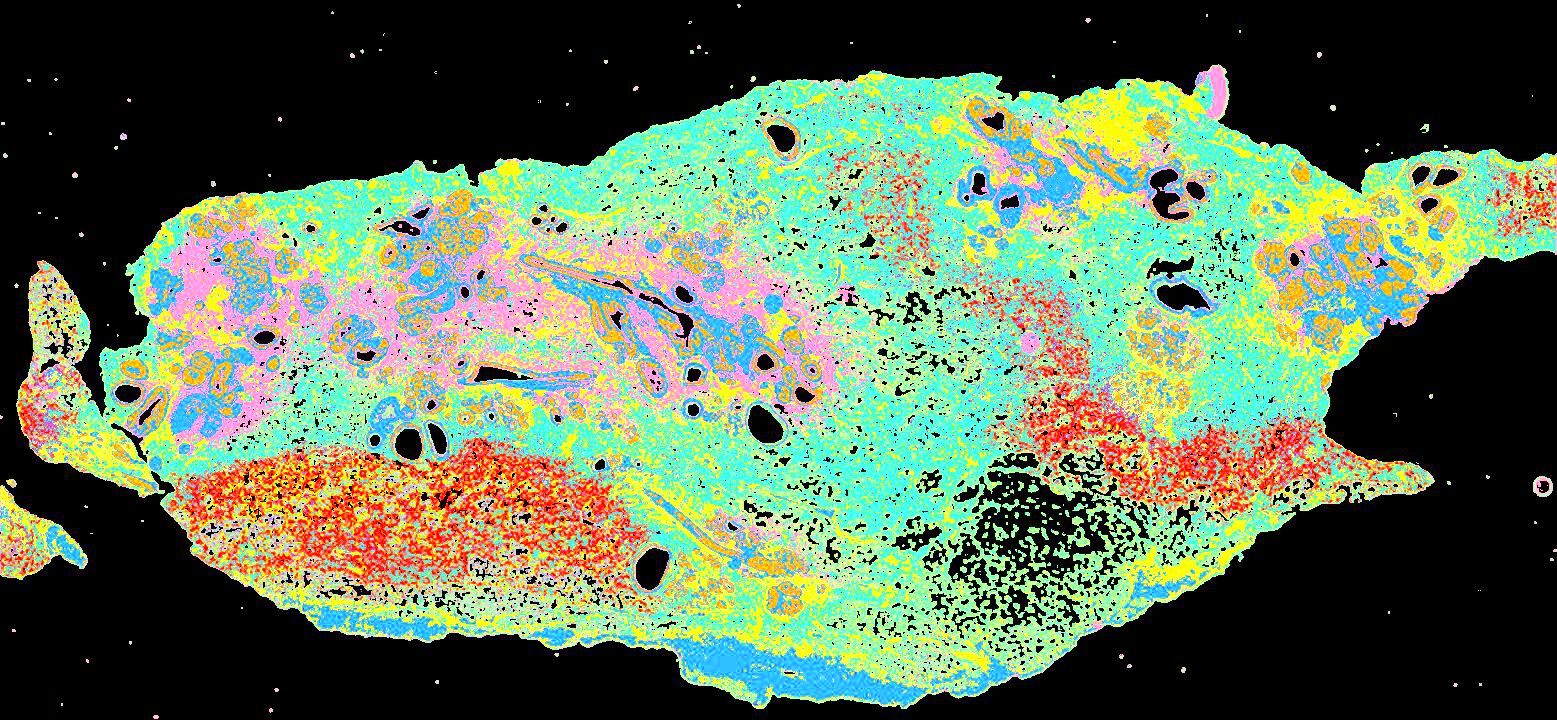
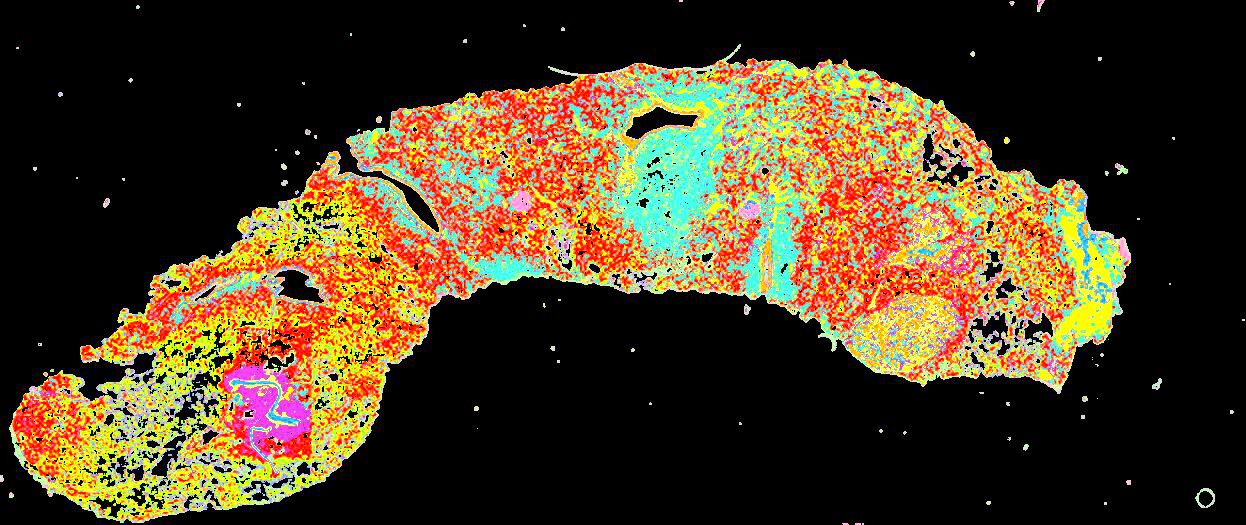
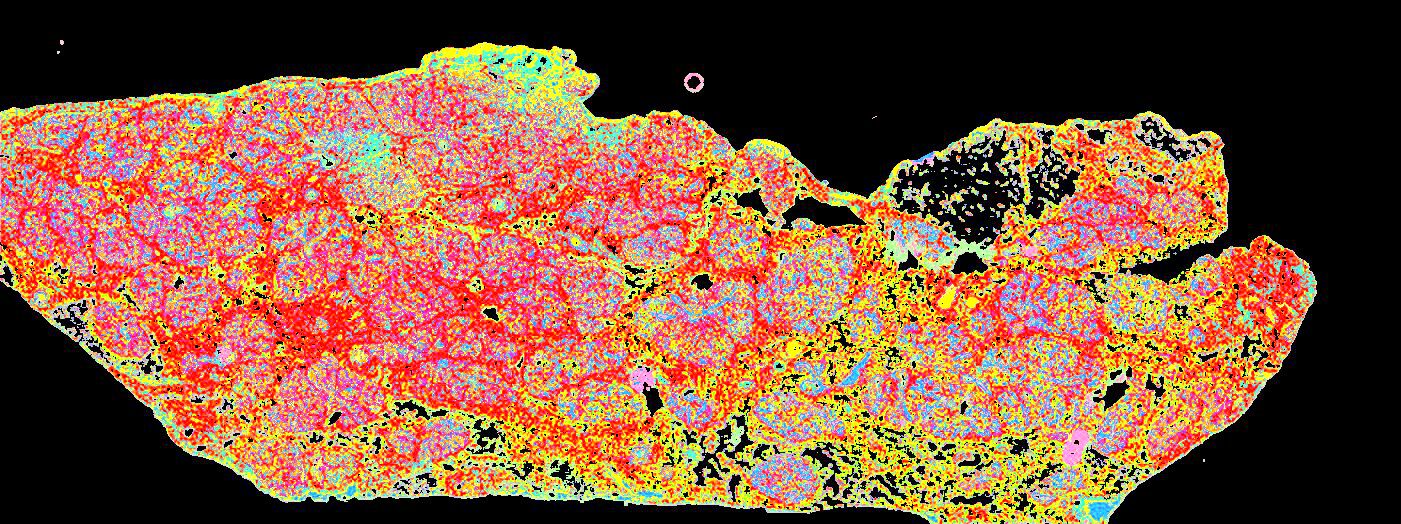
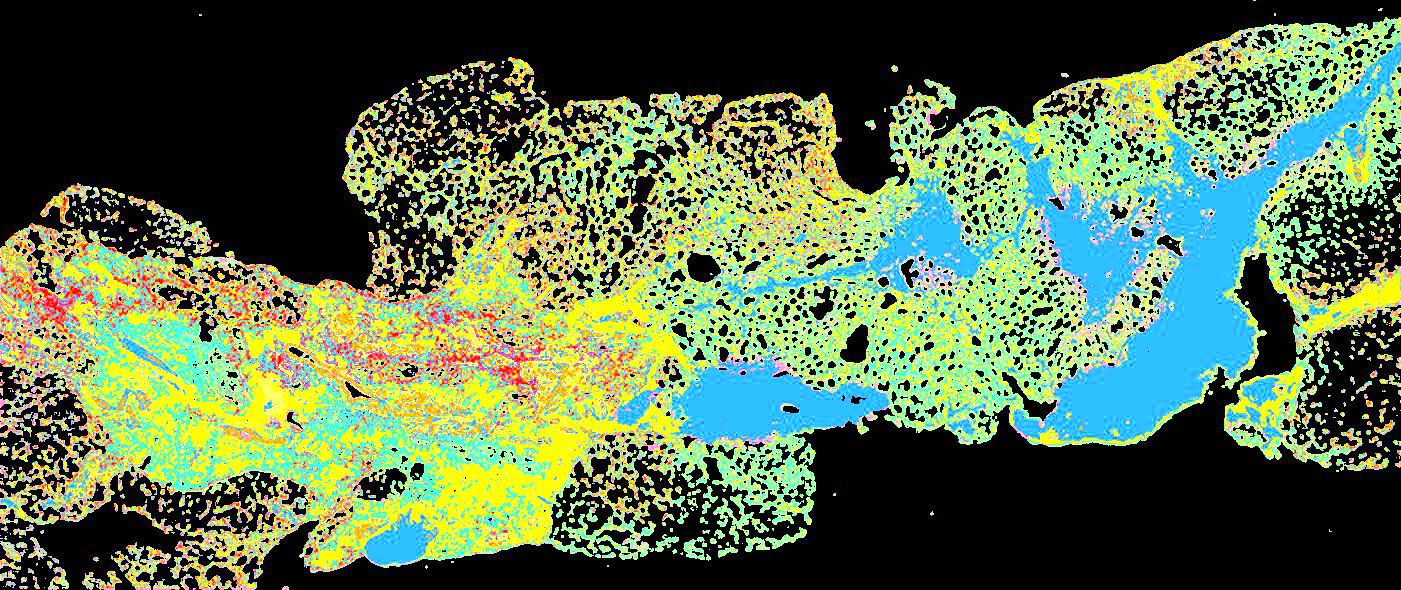
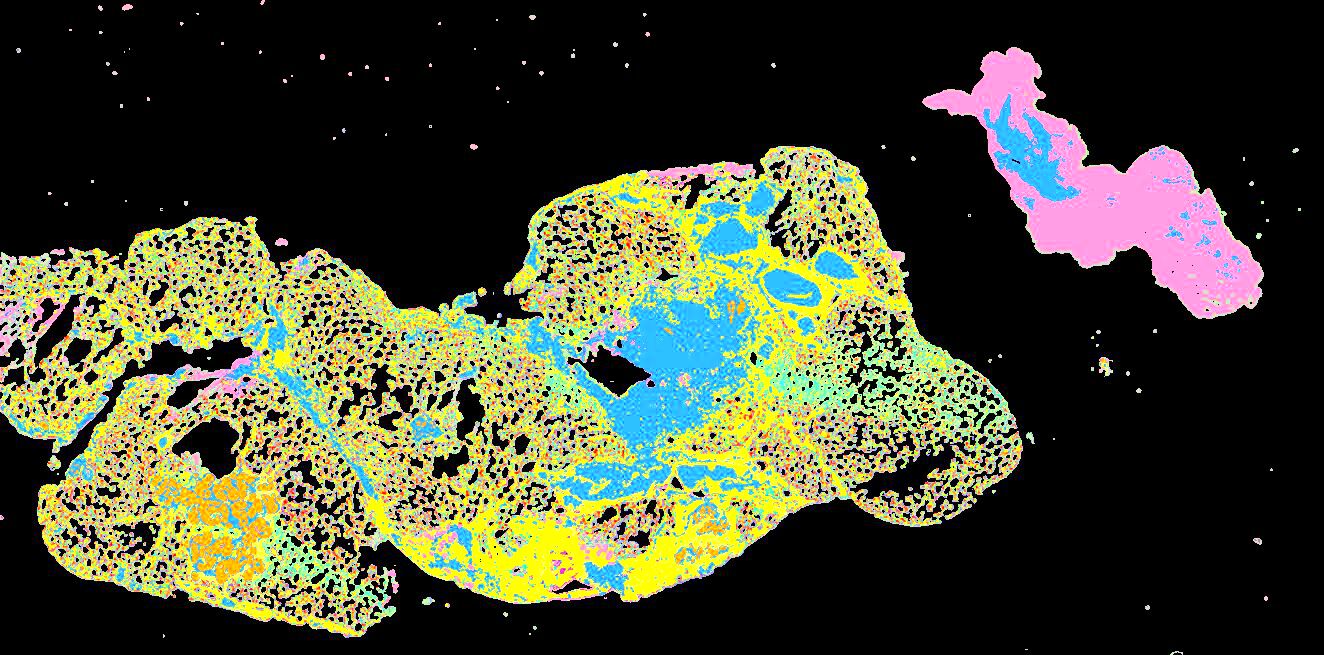
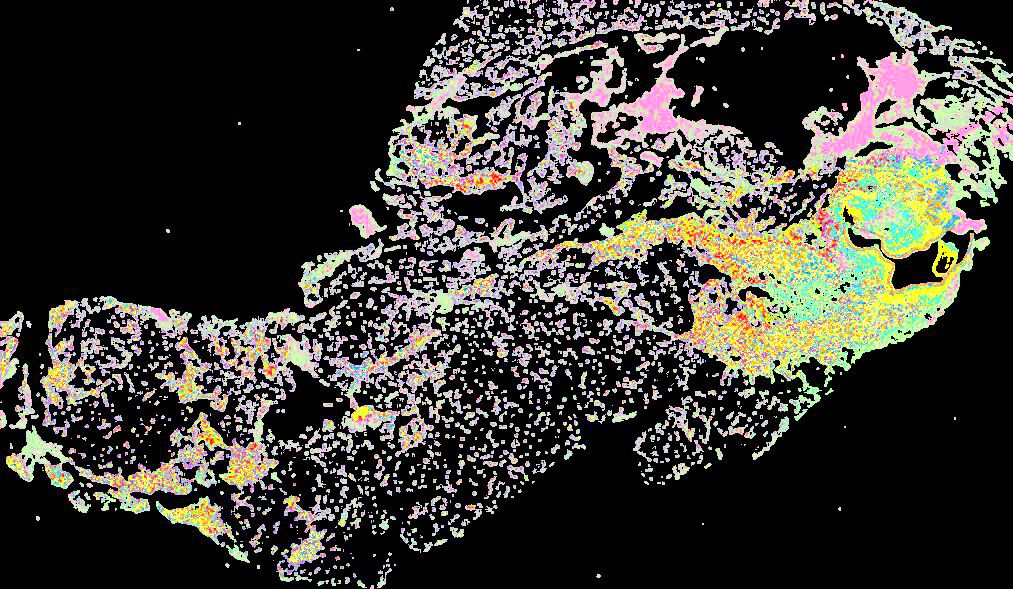
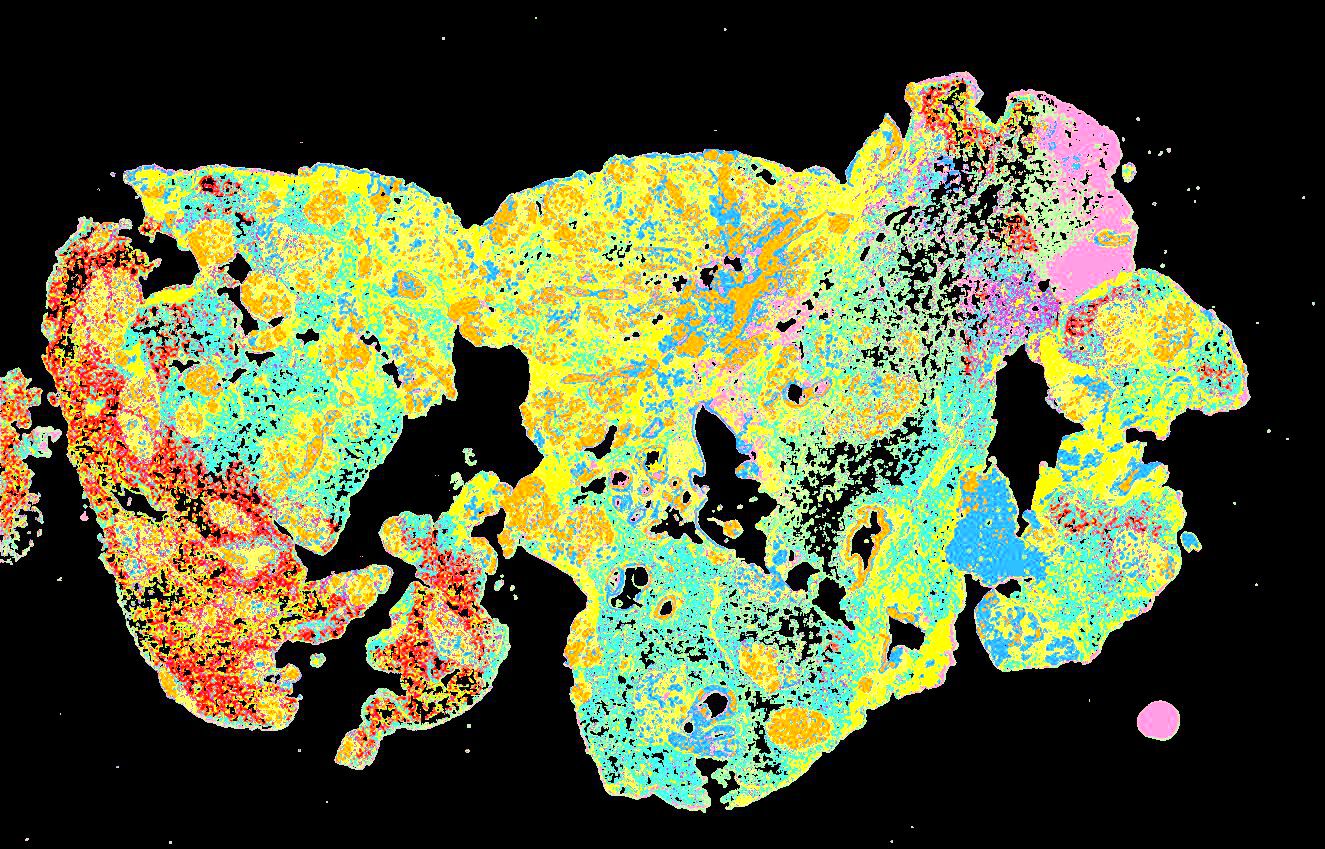
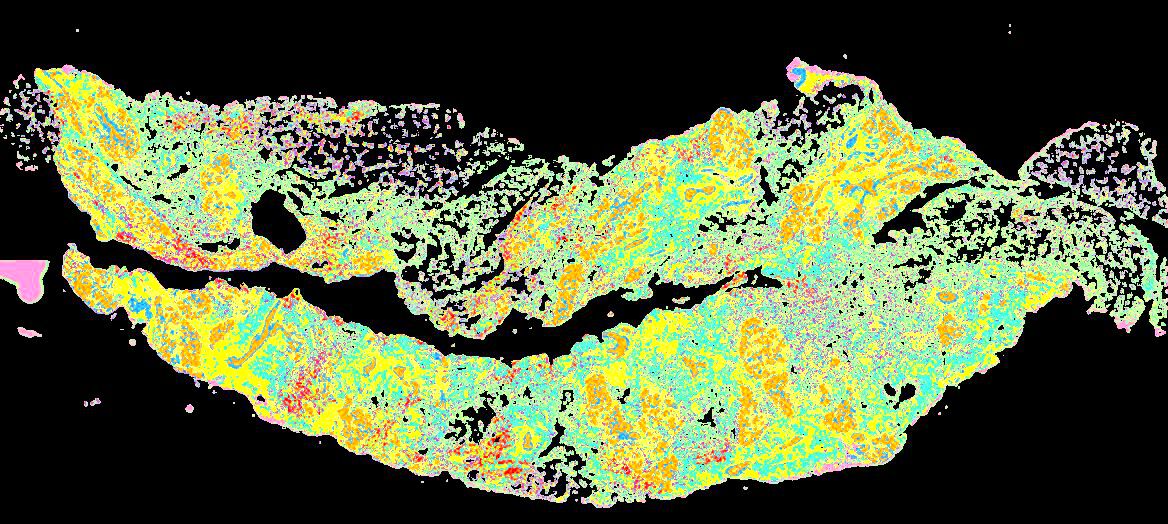
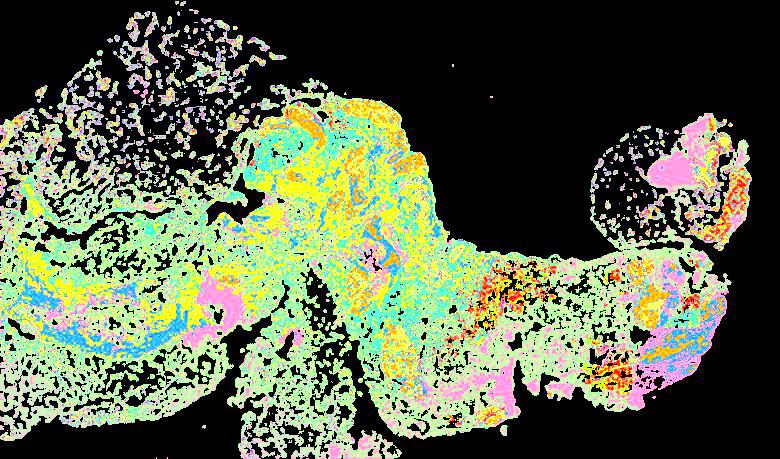
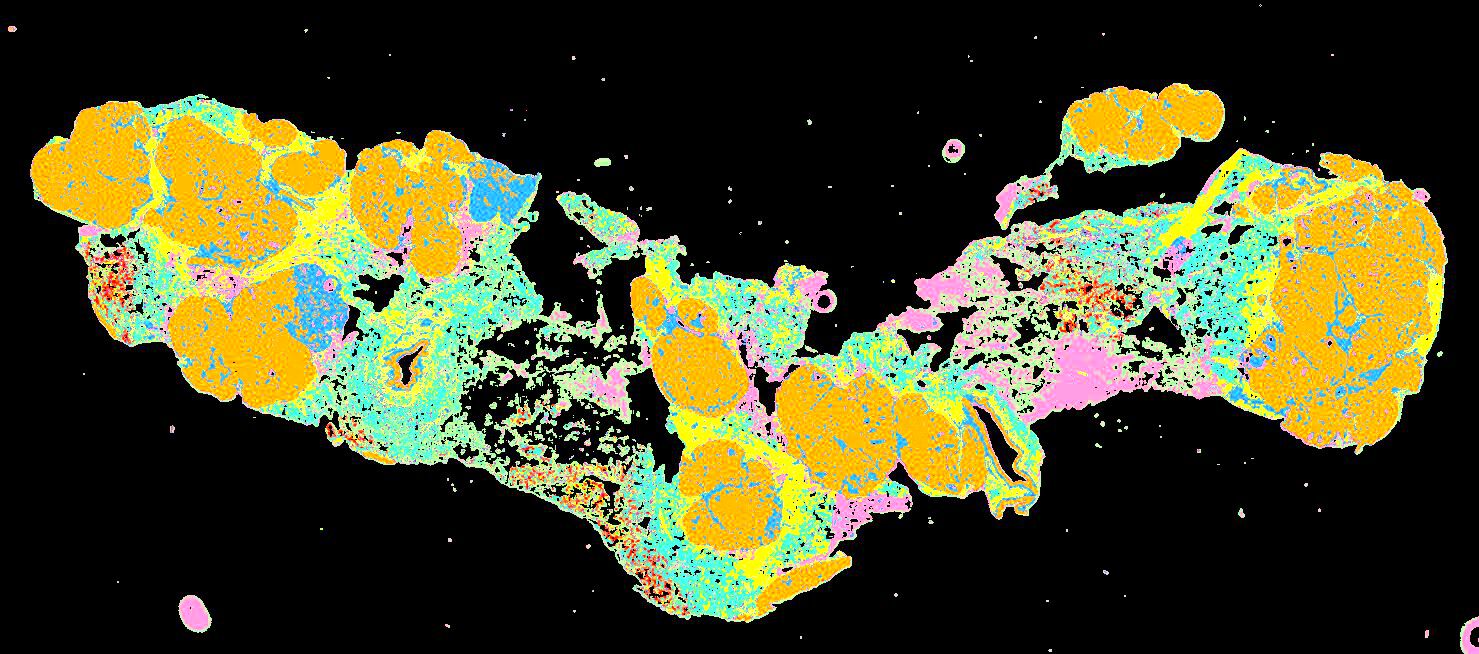
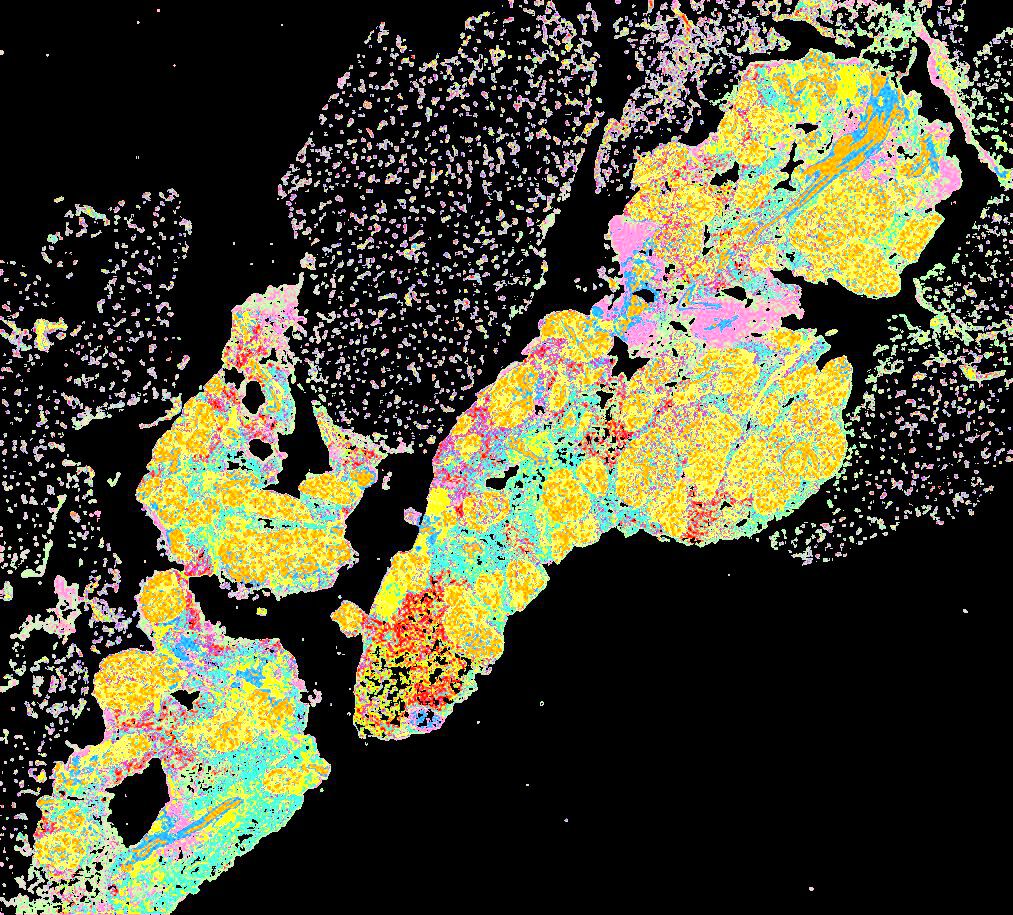
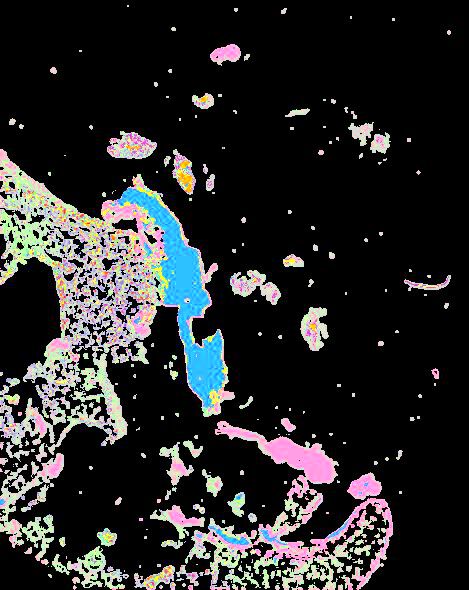
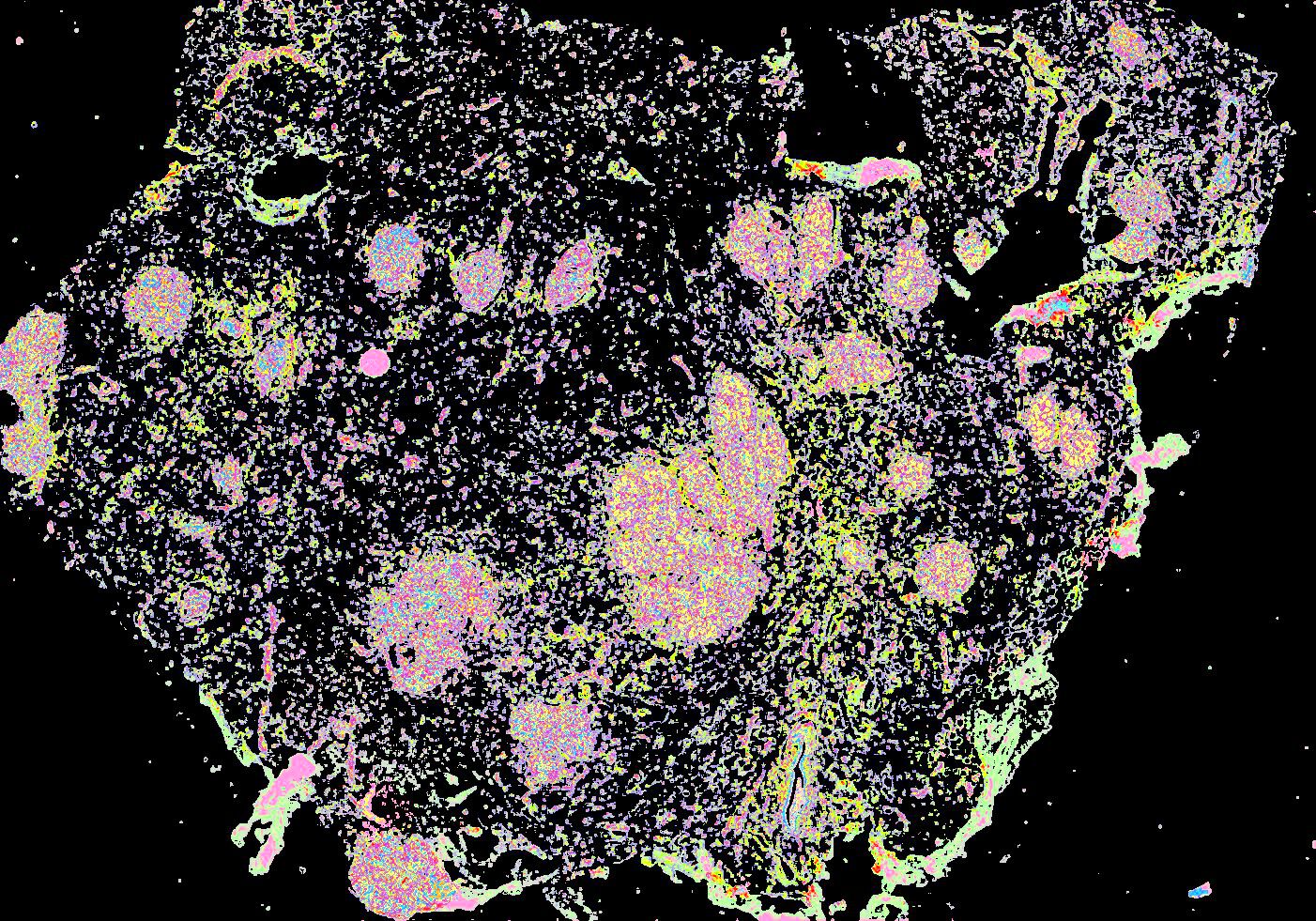
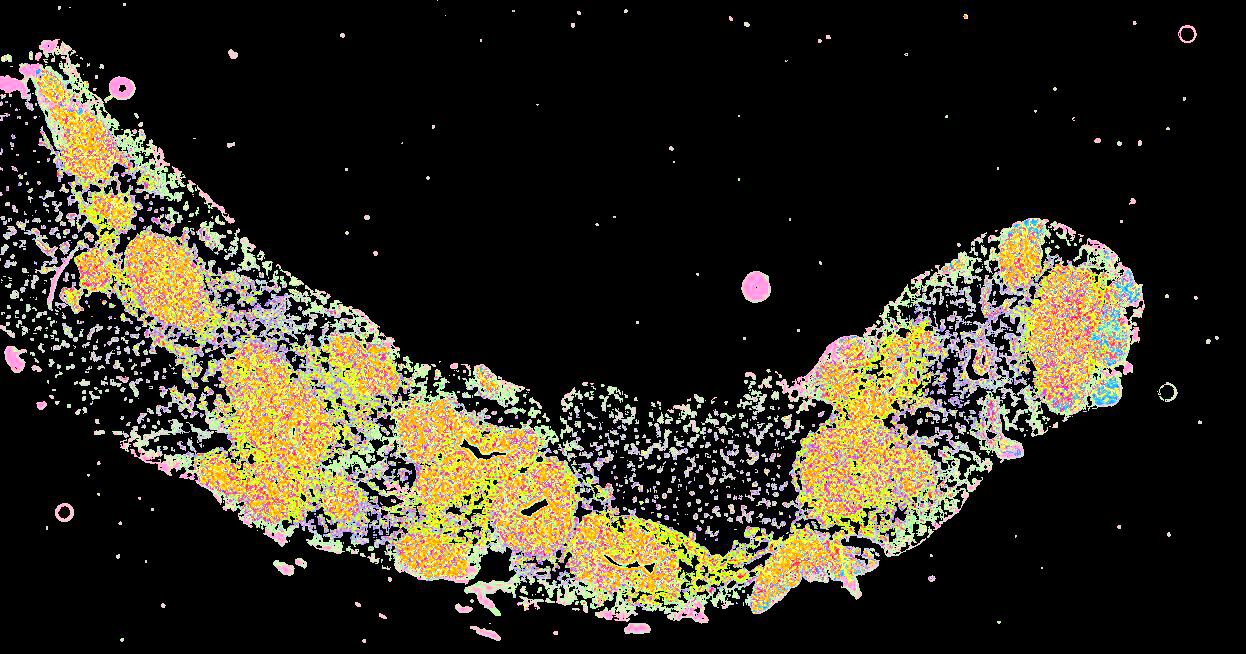
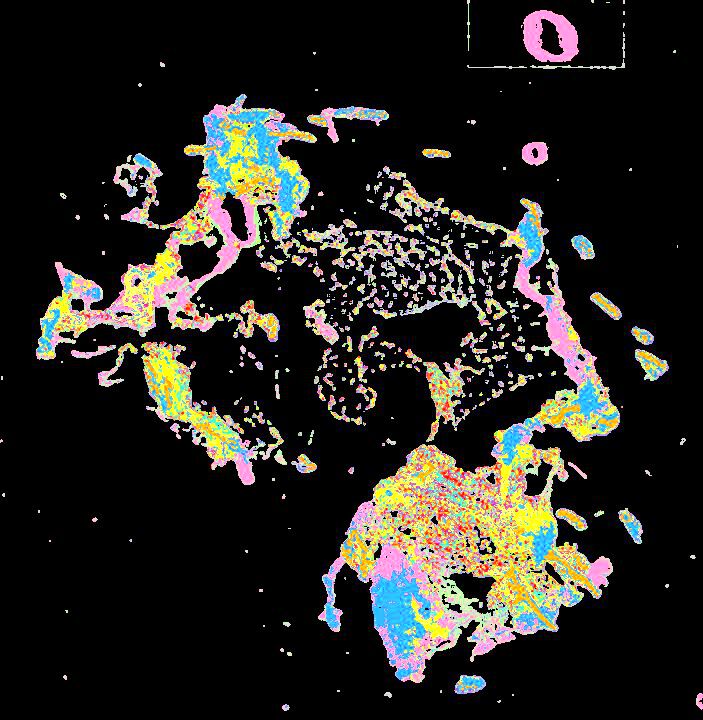
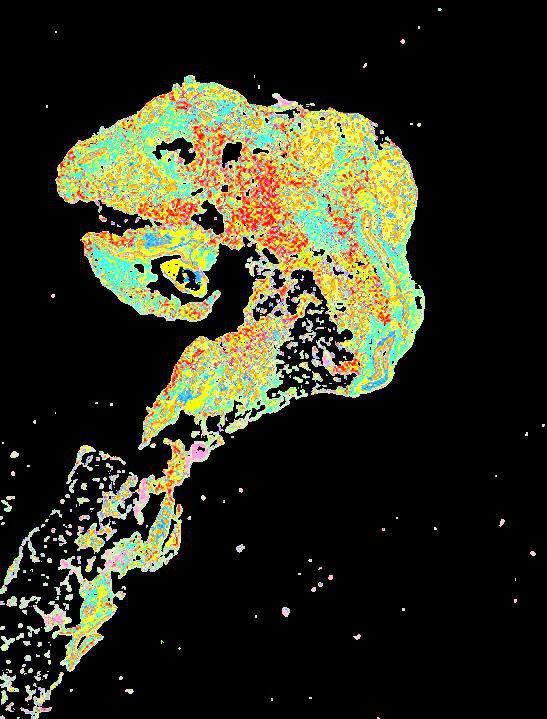
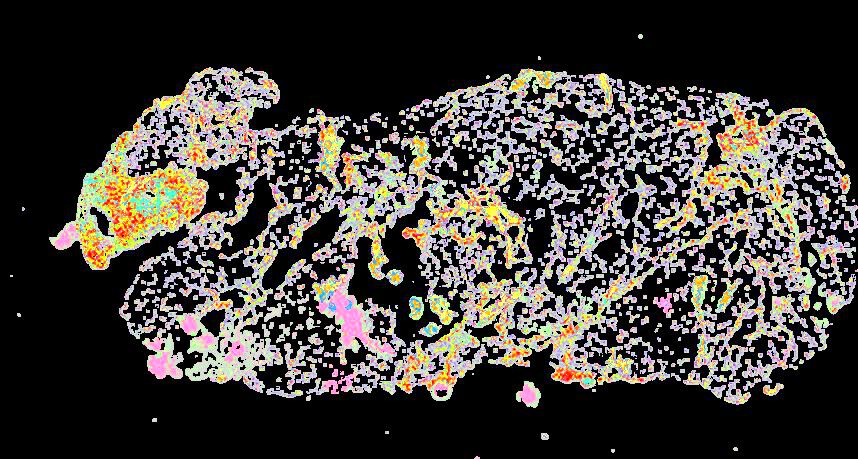
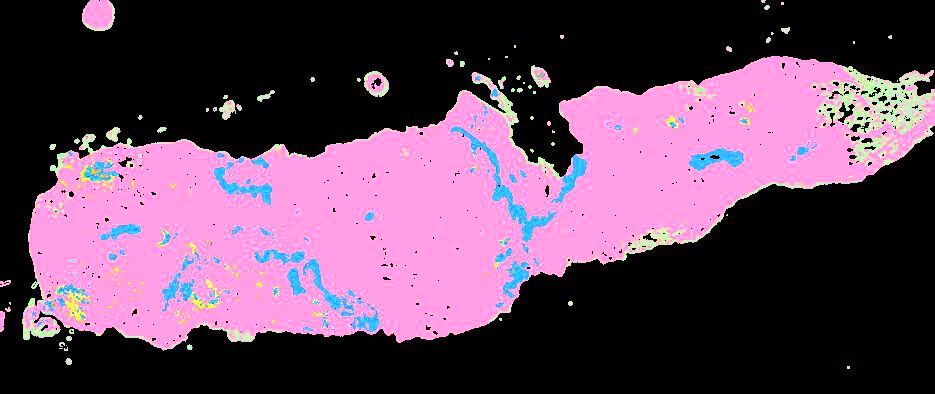
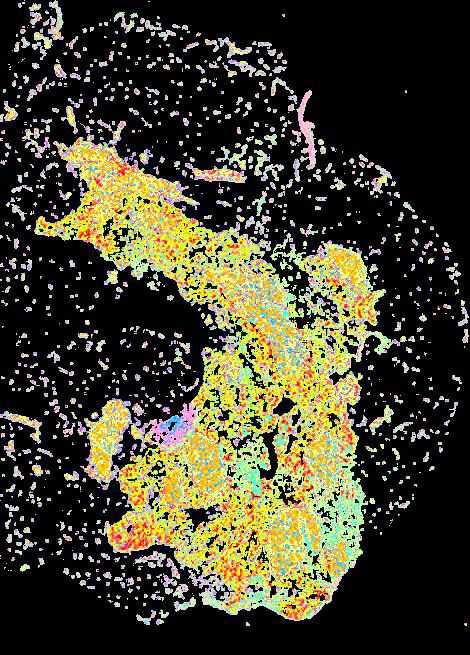
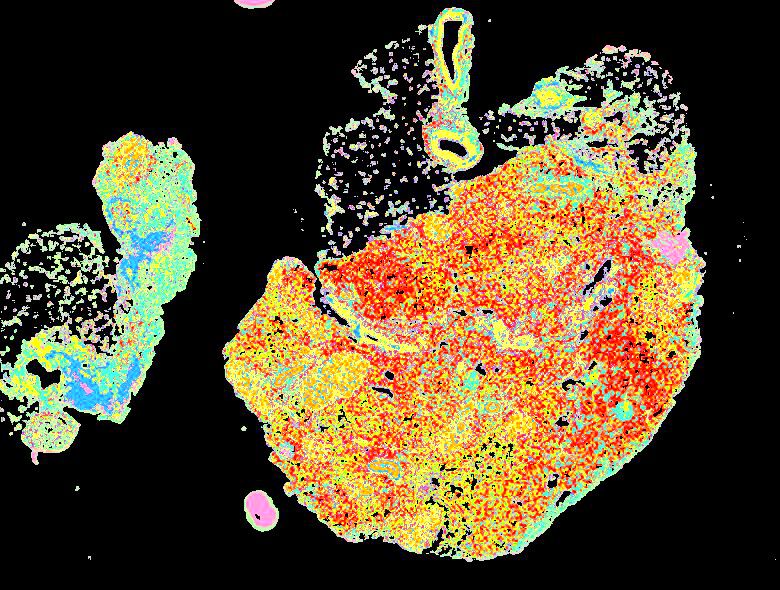
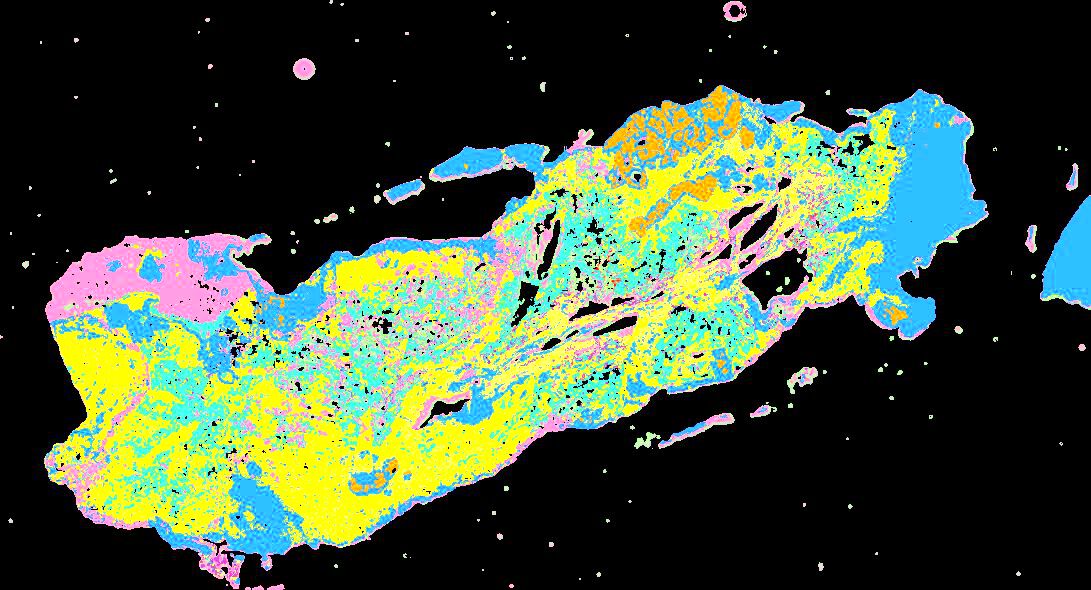
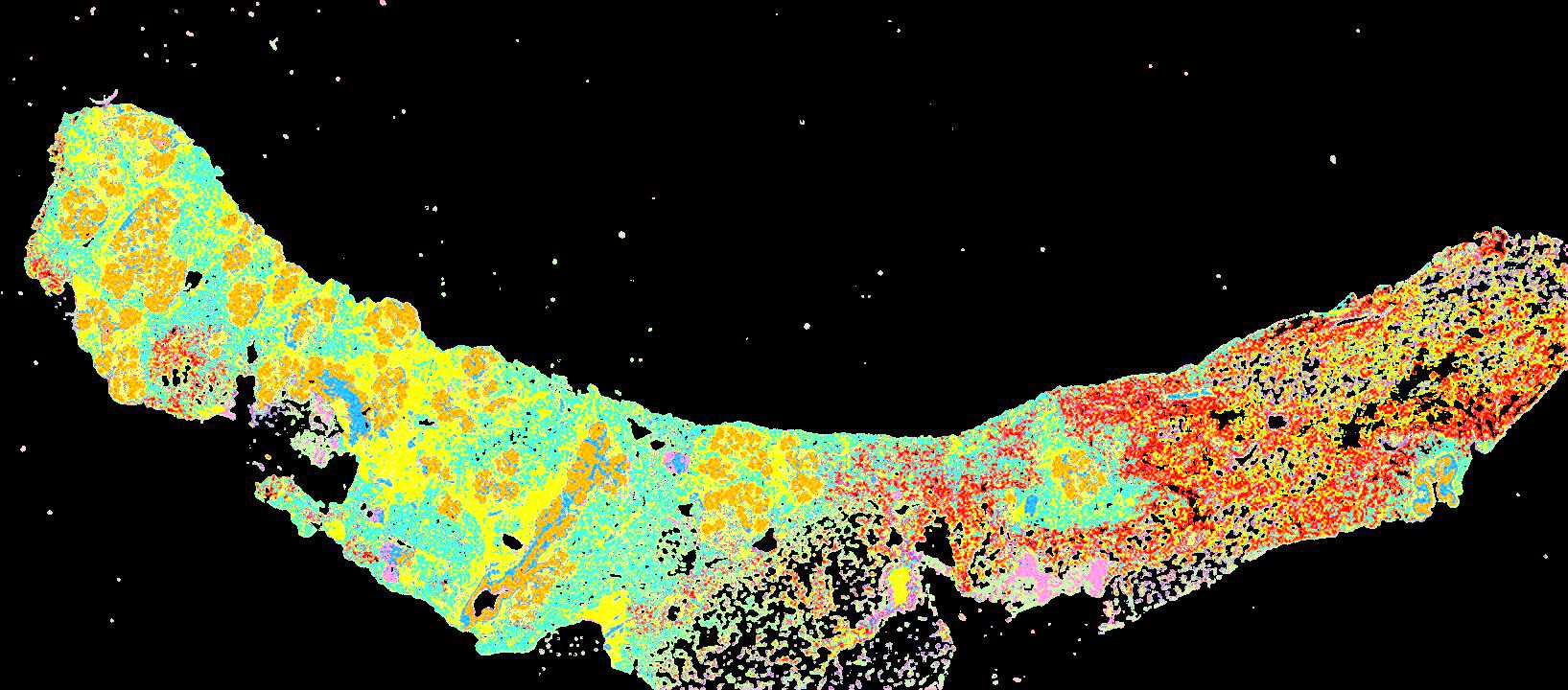
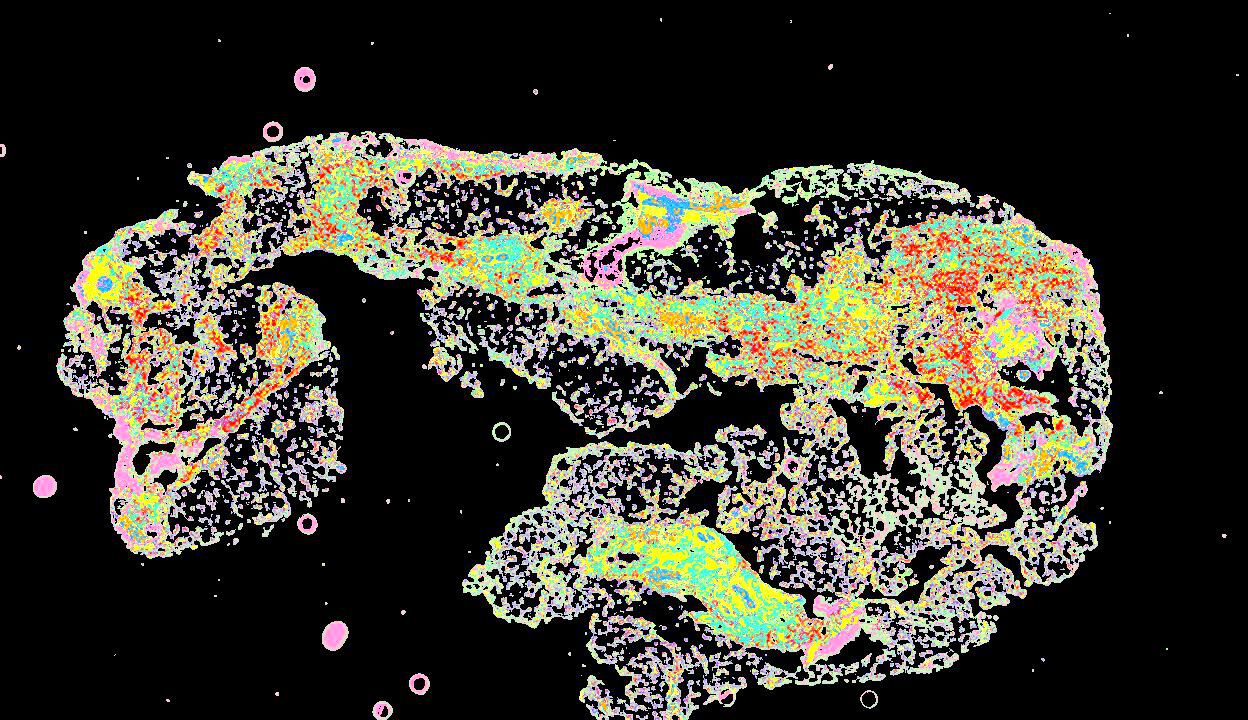
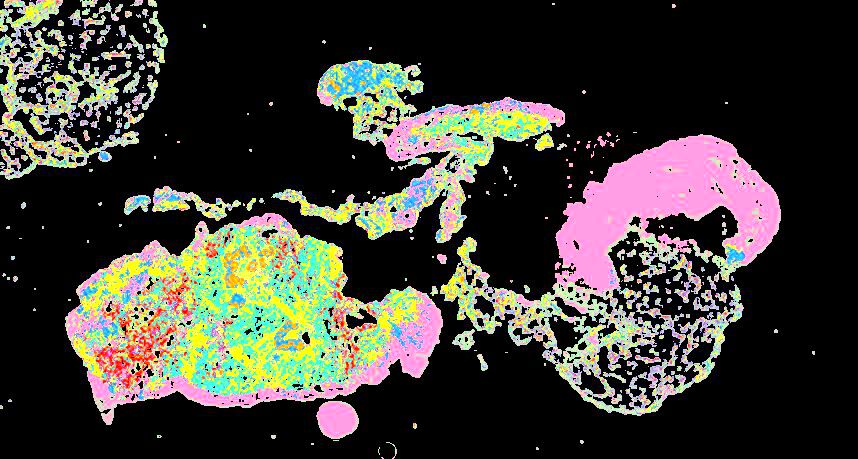
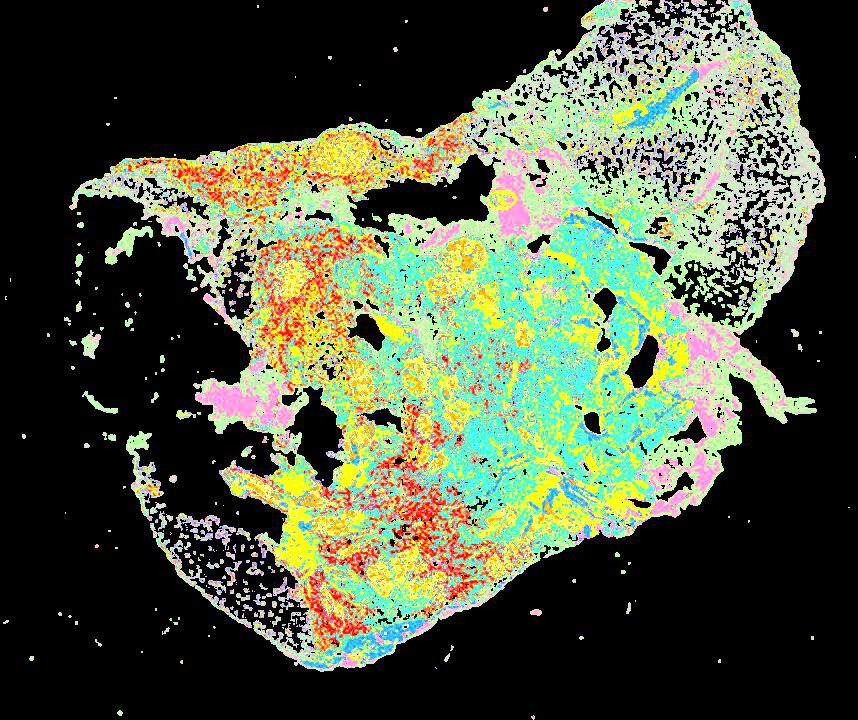
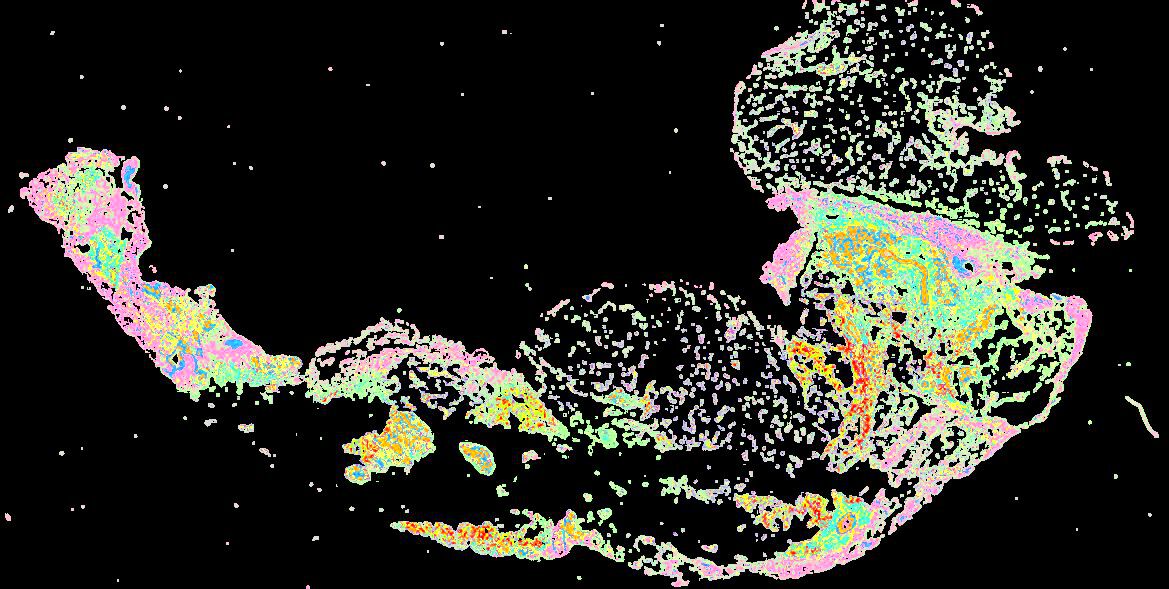
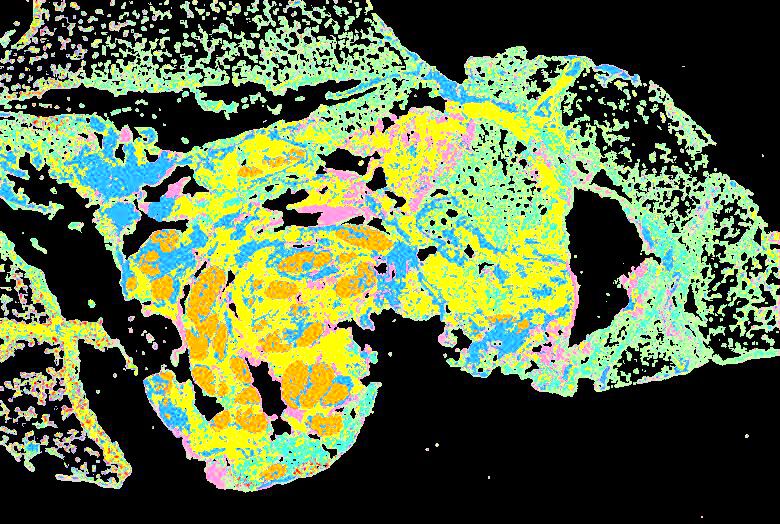
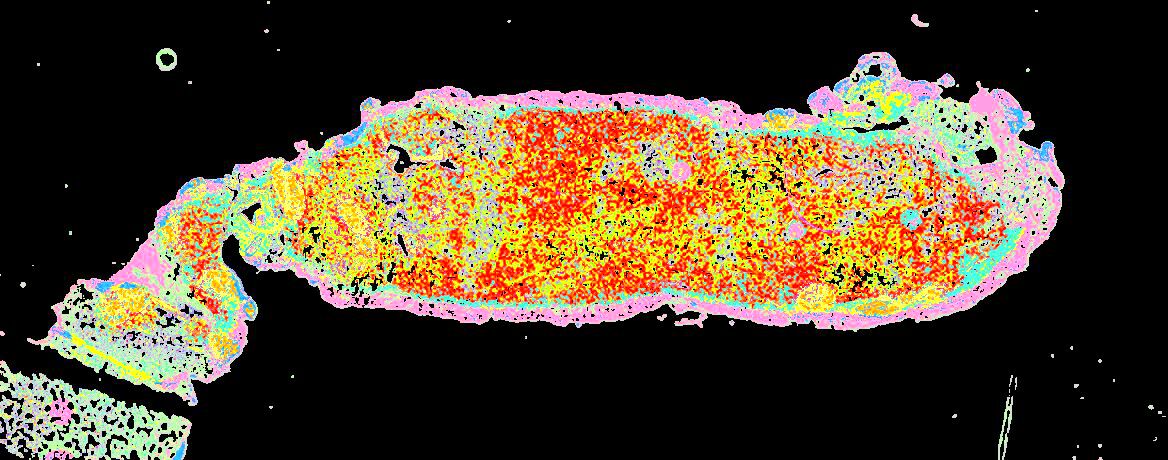
Curated stories provide access to images that have undergone a quality control step to remove failed markers, ensure appropriate channel intensity settings, and provide metadata about the underlying sample and image. Click the Minerva story icon for an interactive view of the full-resolution images.
 Click to enlarge.
Click to enlarge.